2KL3
 
 | | Solution NMR structure of the Rhodanese-like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437A | | Descriptor: | Alr3790 protein | | Authors: | Eletsky, A, Belote, R.L, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Rhodanese-like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437A
To be Published
|
|
1BOI
 
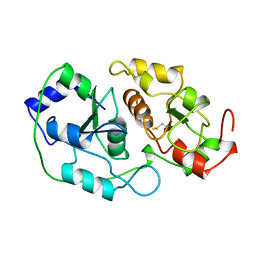 | | N-TERMINALLY TRUNCATED RHODANESE | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1DP2
 
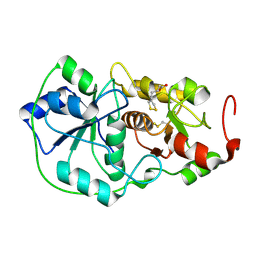 | |
1E0C
 
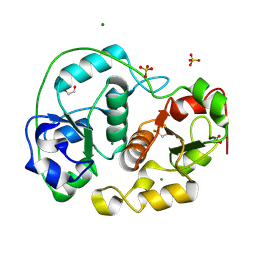 | | SULFURTRANSFERASE FROM AZOTOBACTER VINELANDII | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bordo, D, Deriu, D, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-08 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of a Sulfurtransferase from Azotobacter Vinelandii Highlights the Evolutionary Relationship between the Rhodanese and Phosphatase Enzyme Families
J.Mol.Biol., 298, 2000
|
|
1C25
 
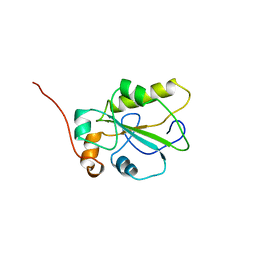 | | HUMAN CDC25A CATALYTIC DOMAIN | | Descriptor: | CDC25A | | Authors: | Fauman, E.B, Cogswell, J.P, Lovejoy, B, Rocque, W.J, Holmes, W, Montana, V.G, Piwnica-Worms, H, Rink, M.J, Saper, M.A. | | Deposit date: | 1998-04-17 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A.
Cell(Cambridge,Mass.), 93, 1998
|
|
1CWT
 
 | | HUMAN CDC25B CATALYTIC DOMAIN WITH METHYL MERCURY | | Descriptor: | CDC25 B-TYPE TYROSINE PHOSPHATASE, CHLORIDE ION, METHYL MERCURY ION, ... | | Authors: | Watenpaugh, K.D, Reynolds, R.A. | | Deposit date: | 1999-08-26 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic subunit of Cdc25B required for G2/M phase transition of the cell cycle.
J.Mol.Biol., 293, 1999
|
|
1CWR
 
 | |
1BOH
 
 | | SULFUR-SUBSTITUTED RHODANESE (ORTHORHOMBIC FORM) | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1CWS
 
 | | HUMAN CDC25B CATALYTIC DOMAIN WITH TUNGSTATE | | Descriptor: | BETA-MERCAPTOETHANOL, CDC25 B-TYPE TYROSINE PHOSPHATASE, CHLORIDE ION, ... | | Authors: | Watenpaugh, K.D, Reynolds, R.A, Chidester, C.G. | | Deposit date: | 1999-08-26 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic subunit of Cdc25B required for G2/M phase transition of the cell cycle.
J.Mol.Biol., 293, 1999
|
|
6K6R
 
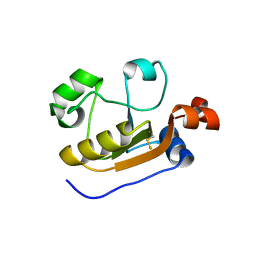 | |
6KTW
 
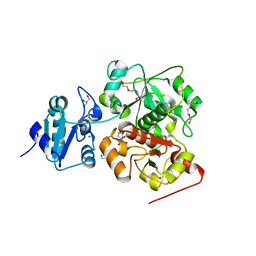 | | structure of EanB with hercynine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KU2
 
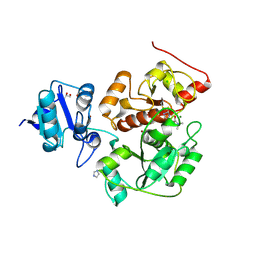 | | The structure of EanB/Y353A complex with ergothioneine covalent linked with persulfide Cys412 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KTZ
 
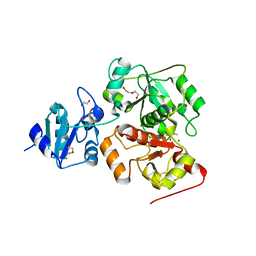 | | The complex structure of EanB/C412S with hercynine | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KTV
 
 | | The structure of EanB complex with hercynine and persulfided Cys412 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KVY
 
 | | The structure of EanB/T414A complex with hercynine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N,N,N-trimethyl-histidine, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of EanB/T414A complex with hercynine
To Be Published
|
|
6KW0
 
 | |
6KTX
 
 | | The wildtype structure of EanB | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KVZ
 
 | |
6KU1
 
 | | The structure of EanB/Y353A complex with ergothioneine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KVW
 
 | |
6MXV
 
 | | The crystal structure of a rhodanese-like family protein from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, ... | | Authors: | Tan, K, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The crystal structure of a rhodanese-like family protein from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
3F4A
 
 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
3HIX
 
 | | Crystal Structure of the Rhodanese_3 like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437i | | Descriptor: | Alr3790 protein, MANGANESE (II) ION | | Authors: | Vorobiev, S, Chen, Y, Forouhar, F, Maglaqui, M, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of the Rhodanese_3 like domain from Anabaena sp Alr3790 protein.
To be Published
|
|
3FS5
 
 | |
3FNJ
 
 | | Crystal structure of the full-length lp_1913 protein from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR140 | | Descriptor: | protein lp_1913 | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-25 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the full-length lp_1913 protein from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR140
To be Published
|
|
