2Y2C
 
 | | crystal structure of AmpD Apoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2B
 
 | | crystal structure of AmpD in complex with reaction products | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2D
 
 | | crystal structure of AmpD holoenzyme | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y28
 
 | | crystal structure of Se-Met AmpD derivative | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
3CLA
 
 | |
2VJ0
 
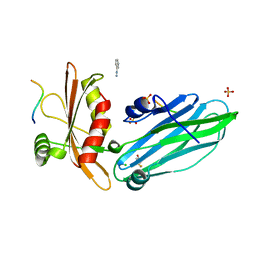 | | Crystal structure of the alpha-adaptin appendage domain, from the AP2 adaptor complex, in complex with an FXDNF peptide from amphiphysin1 and a WVXF peptide from synaptojanin P170 | | Descriptor: | AMPHIPHYSIN, AP-2 COMPLEX SUBUNIT ALPHA-2, BENZAMIDINE, ... | | Authors: | Ford, M.G.J, Praefcke, G.J.K, McMahon, H.T. | | Deposit date: | 2007-12-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Solitary and Repetitive Binding Motifs for the Ap2 Complex {Alpha}-Appendage in Amphiphysin and Other Accessory Proteins.
J.Biol.Chem., 283, 2008
|
|
3U9F
 
 | |
1CLA
 
 | |
2W5P
 
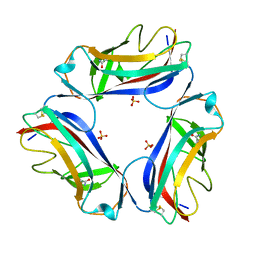 | | DraE Adhesin in complex with Chloramphenicol Succinate (monoclinic form) | | Descriptor: | CHLORAMPHENICOL SUCCINATE, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, SULFATE ION | | Authors: | Pettigrew, D.M, Roversi, P, Davies, S.G, Russell, A.J, Lea, S.M. | | Deposit date: | 2008-12-11 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural Study of the Interaction between the Dr Haemagglutinin Drae and Derivatives of Chloramphenicol
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4PO4
 
 | |
7SHI
 
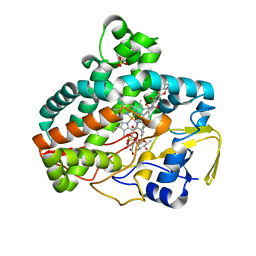 | |
7TJC
 
 | | VHH Chl-B2 in complex with Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, GLYCEROL, VHH-Chl-B2 | | Authors: | Nordeen, S.A, Schwartz, T.U. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and specificity of an anti-chloramphenicol single domain antibody for detection of amphenicol residues.
Protein Sci., 31, 2022
|
|
3GM0
 
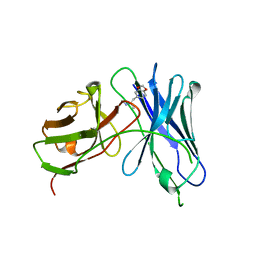 | | Anti-methamphetamine single chain Fv in complex with MDMA | | Descriptor: | (2S)-1-(1,3-benzodioxol-5-yl)-N-methylpropan-2-amine, anti-methamphetamine single chain Fv | | Authors: | Celikel, R, Peterson, E.C, Owens, M, Varughese, K.I. | | Deposit date: | 2009-03-12 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a therapeutic single chain antibody in complex with two drugs of abuse-Methamphetamine and 3,4-methylenedioxymethamphetamine.
Protein Sci., 18, 2009
|
|
1C01
 
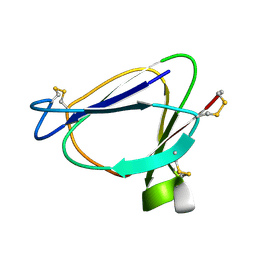 | | SOLUTION STRUCTURE OF MIAMP1, A PLANT ANTIMICROBIAL PROTEIN | | Descriptor: | ANTIMICROBIAL PEPTIDE 1 | | Authors: | McManus, A.M, Nielsen, K.J, Marcus, J.P, Harrison, S.J, Green, J.L, Manners, J.M, Craik, D.J. | | Deposit date: | 1999-07-13 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | MiAMP1, a novel protein from Macadamia integrifolia adopts a Greek key beta-barrel fold unique amongst plant antimicrobial proteins.
J.Mol.Biol., 293, 1999
|
|
6IEY
 
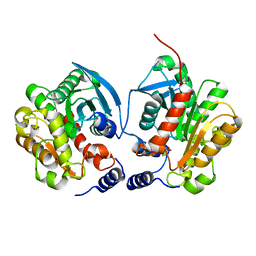 | | Crystal structure of Chloramphenicol-Metabolizaing Enzyme EstDL136-Chloramphenicol complex | | Descriptor: | CHLORAMPHENICOL, Esterase | | Authors: | Kim, S.H, Kang, P.A, Han, K.T, Lee, S.W, Rhee, S.K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome.
PLoS ONE, 14, 2019
|
|
2V92
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2V8Q
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2V9J
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with Mg.ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-23 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
4REW
 
 | | Crystal structure of the non-phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
4RER
 
 | | Crystal structure of the phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex bound to AMP and cyclodextrin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.047 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
4QFR
 
 | | Structure of AMPK in complex with Cl-A769662 activator and STAUROSPORINE inhibitor | | Descriptor: | 2-chloro-4-hydroxy-3-(2'-hydroxybiphenyl-4-yl)-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
4QFS
 
 | | Structure of AMPK in complex with Br2-A769662core activator and STAUROSPORINE inhibitor | | Descriptor: | 2-bromo-3-(4-bromophenyl)-4-hydroxy-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
4QFG
 
 | |
2UV4
 
 | | Crystal Structure of a CBS domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ADENOSINE MONOPHOSPHATE | | Authors: | Day, P, Sharff, A, PArra, L, Cleasby, A, Williams, M, Horer, S, Nar, H, Redemann, N, Tickle, I, Yon, J. | | Deposit date: | 2007-03-09 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure of a Cbs-Domain Pair from the Regulatory Gamma1 Subunit of Human Ampk in Complex with AMP and Zmp.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
