4W84
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4W9D
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methyloxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9O
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4W9Z
 
 | |
4W6V
 
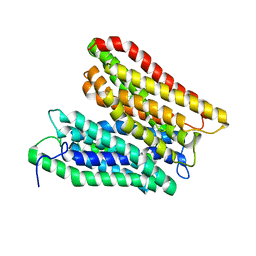 | | Crystal structure of a peptide transporter from Yersinia enterocolitica at 3 A resolution | | Descriptor: | Di-/tripeptide transporter | | Authors: | Jeckelmann, J.-M, Boggavarapu, R, Harder, D, Ucurum, Z, Fotiadis, D. | | Deposit date: | 2014-08-21 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.01819 Å) | | Cite: | Role of electrostatic interactions for ligand recognition and specificity of peptide transporters.
Bmc Biol., 13, 2015
|
|
3MQO
 
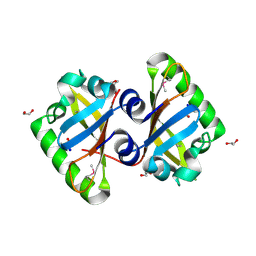 | | The Crystal Structure of the PAS domain in complex with isopropanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.7A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Stein, A.J, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the PAS domain in complex with isopropanol of a Transcriptional Regulator in the LuxR family from Burkholderia thailandensis to 1.7A
To be Published
|
|
2RGC
 
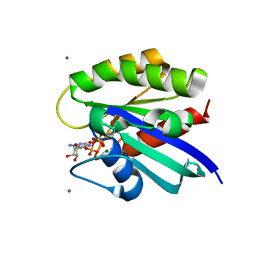 | | Crystal structure of H-RasQ61V-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Buhrman, G, Wink, G, Mattos, C. | | Deposit date: | 2007-10-03 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Transformation Efficiency of RasQ61 Mutants Linked to Structural Features of the Switch Regions in the Presence of Raf.
Structure, 15, 2007
|
|
4W7T
 
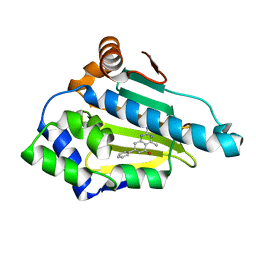 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
2RI4
 
 | |
2RJ2
 
 | | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution | | Descriptor: | CHLORIDE ION, F-box only protein 2, NICKEL (II) ION | | Authors: | Vaijayanthimala, S, Velmurugan, D, Mizushima, T, Yamane, T, Yoshida, Y, Tanaka, K. | | Deposit date: | 2007-10-14 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution
To be Published
|
|
4W9G
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(3-methyl-4-(thiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[3-methyl-4-(1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9N
 
 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase complexed with triclosan | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, IMIDAZOLE, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
2RGA
 
 | | Crystal structure of H-RasQ61I-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Buhrman, G, Wink, G, Mattos, C. | | Deposit date: | 2007-10-03 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transformation Efficiency of RasQ61 Mutants Linked to Structural Features of the Switch Regions in the Presence of Raf.
Structure, 15, 2007
|
|
4WAV
 
 | | Crystal Structure of Haloquadratum walsbyi bacteriorhodopsin mutant D93N | | Descriptor: | Bacteriorhodopsin-I, RETINAL, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Wang, A.H.J, Hsu, M.F, Yang, C.S, Fu, H.Y. | | Deposit date: | 2014-09-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an acid-tolerant light-driven proton pump at 1.85 Angstroms resolution
To be published
|
|
3LZJ
 
 | | RB69 DNA Polymerase (Y567A) ternary complex with dCTP Opposite 7,8-Dihydro-8-oxoguanine | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-03-01 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
2RHD
 
 | | Crystal structure of Cryptosporidium parvum small GTPase RAB1A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Small GTP binding protein rab1a | | Authors: | Dong, A, Xu, X, Lew, J, Lin, Y.H, Khuu, C, Sun, X, Qiu, W, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of Cryptosporidium parvum small GTPase RAB1A.
To be Published
|
|
2RN1
 
 | | Liquid crystal solution structure of the kissing complex formed by the apical loop of the HIV TAR RNA and a high affinity RNA aptamer optimized by SELEX | | Descriptor: | RNA (5'-R(P*GP*AP*GP*CP*CP*CP*UP*GP*GP*GP*AP*GP*GP*CP*UP*C)-3'), RNA (5'-R(P*GP*CP*UP*GP*GP*UP*CP*CP*CP*AP*GP*AP*CP*AP*GP*C)-3') | | Authors: | Van Melckebeke, H, Devany, M, Di Primo, C, Beaurain, F, Toulme, J, Bryce, D.L, Boisbouvier, J. | | Deposit date: | 2007-12-05 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Liquid-crystal NMR structure of HIV TAR RNA bound to its SELEX RNA aptamer reveals the origins of the high stability of the complex
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4WC5
 
 | | Structure of tRNA-processing enzyme complex 3 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Poly A polymerase, RNA (74-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4WE7
 
 | | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
2RVE
 
 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*CP*TP*CP*G)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilson, K.S. | | Deposit date: | 1991-03-19 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
2RVO
 
 | | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish | | Descriptor: | RNA (34-MER) | | Authors: | Otsu, M, Norose, N, Arai, N, Terao, R, Kajikawa, M, Okada, N, Kawai, G. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish.
J. Biochem., 162, 2017
|
|
2RT8
 
 | | Structure of metallo-dna in solution | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | Deposit date: | 2013-06-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
2RSK
 
 | | RNA aptamer against prion protein in complex with the partial binding peptide | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3'), partial binding peptide of Major prion protein | | Authors: | Mashima, T, Nishikawa, F, Kamatari, Y.O, Fujiwara, H, Nishikawa, S, Kuwata, K, Katahira, M. | | Deposit date: | 2012-03-08 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Anti-prion activity of an RNA aptamer and its structural basis
Nucleic Acids Res., 41, 2013
|
|
2RFN
 
 | | x-ray structure of c-Met with inhibitor. | | Descriptor: | 2-benzyl-5-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
2RLH
 
 | |
