4IEM
 
 | | Human apurinic/apyrimidinic endonuclease (APE1) with product DNA and Mg2+ | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*C)-3'), DNA (5'-D(P*(3DR)P*GP*AP*TP*CP*G)-3'), ... | | Authors: | Tsutakawa, S.E, Mol, C.D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3936 Å) | | Cite: | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
2Q6N
 
 | | Structure of Cytochrome P450 2B4 with Bound 1-(4-cholorophenyl)imidazole | | Descriptor: | 1-(4-CHLOROPHENYL)-1H-IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhao, Y, Sun, L, Muralidhara, B.K, Kumar, S, White, M.A, Stout, C.D, Halpert, J.R. | | Deposit date: | 2007-06-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4.
Biochemistry, 46, 2007
|
|
4ETZ
 
 | | Crystal Structure of PelD 158-CT from Pseudomonas aeruginosa PAO1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), PelD | | Authors: | Li, Z, Chen, J, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the PelD Cyclic Diguanylate Effector Involved in Pellicle Formation in Pseudomonas aeruginosa PAO1.
J.Biol.Chem., 287, 2012
|
|
4IBD
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-methylbenzoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4EUD
 
 | |
3DQG
 
 | | Peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans. | | Descriptor: | Heat shock 70 kDa protein F | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans.
To be Published
|
|
4EUY
 
 | | Crystal structure of thioredoxin-like protein BCE_0499 from Bacillus cereus ATCC 10987 | | Descriptor: | Uncharacterized protein | | Authors: | Shabalin, I.G, Kagan, O, Chruszcz, M, Grabowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of thioredoxin-like protein BCE_0499 from
Bacillus cereus
To be Published
|
|
3TQT
 
 | | Structure of the D-alanine-D-alanine ligase from Coxiella burnetii | | Descriptor: | D-alanine--D-alanine ligase | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2Q8T
 
 | | Crystal Structure of the CC chemokine CCL14 | | Descriptor: | CCL14 | | Authors: | Blain, K.Y. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Functional Characterization of CC Chemokine CCL14
Biochemistry, 46, 2007
|
|
3DQY
 
 | | Crystal structure of Toluene 2,3-Dioxygenase Ferredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Toluene 1,2-dioxygenase system ferredoxin subunit | | Authors: | Friemann, R, Lee, K, Brown, E.N, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2008-07-10 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4IBW
 
 | | Human p53 core domain with hot spot mutation R273H and second-site suppressor mutation T284R in sequence-specific complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA (5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3'), ... | | Authors: | Eldar, A, Rozenberg, H, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
2Q98
 
 | |
3DE7
 
 | |
3TRB
 
 | | Structure of an addiction module antidote protein of a HigA (higA) family from Coxiella burnetii | | Descriptor: | Virulence-associated protein I | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQN
 
 | | Structure of the transcriptional regulator of the GntR family, from Coxiella burnetii. | | Descriptor: | Transcriptional regulator, GntR family | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQZ
 
 | | Structure of a deoxyuridine 5'-triphosphate nucleotidohydrolase (dut) from Coxiella burnetii | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, SULFATE ION | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3DKK
 
 | | Aged Form of Human Butyrylcholinesterase Inhibited by Tabun | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nachon, F, Carletti, E. | | Deposit date: | 2008-06-25 | | Release date: | 2008-12-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
4EY7
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Donepezil | | Descriptor: | 1,2-ETHANEDIOL, 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheung, J, Rudolph, M, Burshteyn, F, Cassidy, M, Gary, E, Love, J, Height, J, Franklin, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3509 Å) | | Cite: | Structures of human acetylcholinesterase in complex with pharmacologically important ligands.
J.Med.Chem., 55, 2012
|
|
3TR6
 
 | | Structure of a O-methyltransferase from Coxiella burnetii | | Descriptor: | NICKEL (II) ION, O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3DFH
 
 | | crystal structure of putative mandelate racemase / muconate lactonizing enzyme from Vibrionales bacterium SWAT-3 | | Descriptor: | SODIUM ION, mandelate racemase | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | crystal structure of putative mandelate racemase / muconate lactonizing enzyme from Vibrionales bacterium SWAT-3
To be Published
|
|
3TR9
 
 | | Structure of a dihydropteroate synthase (folP) in complex with pteroic acid from Coxiella burnetii | | Descriptor: | CHLORIDE ION, Dihydropteroate synthase, PTEROIC ACID, ... | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3DG1
 
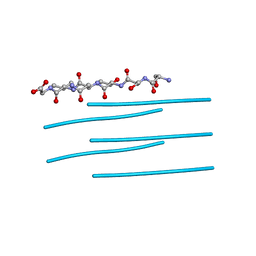 | | Segment SSTNVG derived from IAPP | | Descriptor: | SSTNVG from Islet Amyloid Polypeptide | | Authors: | Wiltzius, J.J, Sievers, S.A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin).
Protein Sci., 17, 2008
|
|
2PY0
 
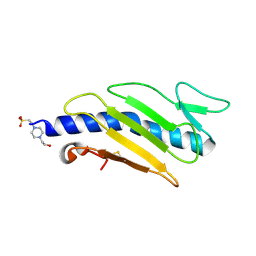 | | Crystal structure of Cs1 pilin chimera | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fimbrial protein | | Authors: | Kao, D.J, Churchill, M.E, Hodges, R.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Animal Protection and Structural Studies of a Consensus Sequence Vaccine Targeting the Receptor Binding Domain of the Type IV Pilus of Pseudomonas aeruginosa.
J.Mol.Biol., 374, 2007
|
|
3TTI
 
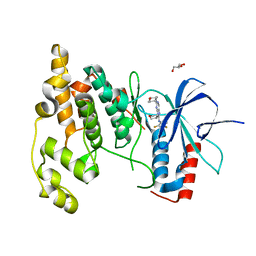 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2PU9
 
 | |
