2MGK
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURES OF FIVE DISTAL HISTIDINE MUTANTS OF SPERM WHALE MYOGLOBIN | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Quillin, M.L, Arduini, R.M, Phillips Jr, G.N. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of distal histidine mutants of sperm whale myoglobin.
J.Mol.Biol., 234, 1993
|
|
2MB5
 
 | |
2MGF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURES OF FIVE DISTAL HISTIDINE MUTANTS OF SPERM WHALE MYOGLOBIN | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Quillin, M.L, Arduini, R.M, Phillips Jr, G.N. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution crystal structures of distal histidine mutants of sperm whale myoglobin.
J.Mol.Biol., 234, 1993
|
|
2HS9
 
 | |
6Z53
 
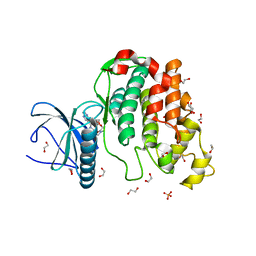 | | Crystal structure of CLK3 in complex with macrocycle ODS2003128 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003128
To Be Published
|
|
2MKS
 
 | |
2HT8
 
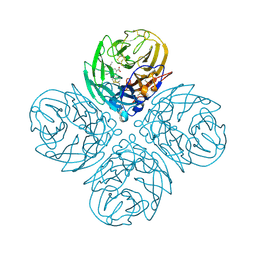 | | N8 neuraminidase in complex with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
6Z83
 
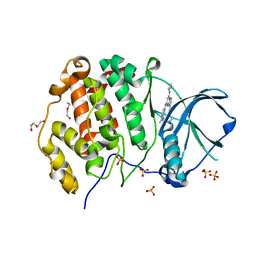 | | CK2 alpha bound to chemical probe SGC-CK2-1 | | Descriptor: | Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
2M3U
 
 | |
2FDV
 
 | | Microsomal P450 2A6 with the inhibitor N-Methyl(5-(pyridin-3-yl)furan-2-yl)methanamine bound | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 2A6, N-METHYL(5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, ... | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
2FJW
 
 | | d(CTTGAATGCATTCAAG) in complex with MMLV RT catalytic fragment | | Descriptor: | 5'-D(*CP*TP*TP*GP*AP*AP*TP*G)-3', 5'-D(P*CP*AP*TP*TP*CP*AP*AP*G)-3', Reverse transcriptase | | Authors: | Goodwin, K.D, Georgiadis, M.M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A High-Throughput, High-Resolution Strategy for the Study of Site-Selective DNA Binding Agents: Analysis of a "Highly Twisted" Benzimidazole-Diamidine.
J.Am.Chem.Soc., 128, 2006
|
|
2FLE
 
 | | Structural analysis of asymmetric inhibitor bound to the HIV-1 Protease V82A mutant | | Descriptor: | (2S,2'S)-N,N'-[(2S,3S,4S,5S)-1-CYCLOHEXYL-3,4-DIHYDROXY-6-PHENYLHEXANE-2,5-DIYL]BIS[3-METHYL-2-({[METHYL(PYRIDIN-2-YLMETHYL)AMINO]CARBONYL}AMINO)BUTANAMIDE], GLYCEROL, pol protein | | Authors: | Clemente, J.C, Robbins, A, Dunn, B.M, Sussman, F. | | Deposit date: | 2006-01-05 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design, synthesis, evaluation, and crystallographic-based structural studies of HIV-1 protease inhibitors with reduced response to the V82A mutation.
J.Med.Chem., 51, 2008
|
|
6YU1
 
 | | CLK3 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
4J1R
 
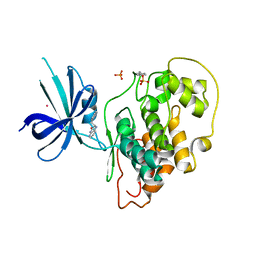 | | Crystal Structure of GSK3b in complex with inhibitor 15R | | Descriptor: | (2R)-2-(1H-indol-3-ylmethyl)-1,4-dihydropyrido[2,3-b]pyrazin-3(2H)-one, Glycogen synthase kinase-3 beta, PHOSPHATE ION, ... | | Authors: | Zhan, C, Wang, Y, Wach, J, Sheehan, P, Zhong, C, Harris, R, Patskovsky, Y, Bishop, J, Haggarty, S, Ramek, A, Berry, K, O'Herin, C, Koehler, A.N, Hung, A.W, Young, D.W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Fragment-based approach using diversity-oriented synthesis yields a GSK3b inhibitor
To be Published
|
|
2FLJ
 
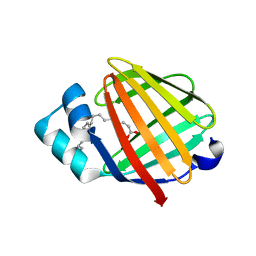 | | Fatty acid binding protein from locust flight muscle in complex with oleate | | Descriptor: | Fatty acid-binding protein, OLEIC ACID | | Authors: | Luecke, C, Qiao, Y, van Moerkerk, H.T.B, Veerkamp, J.H, Hamilton, J.A. | | Deposit date: | 2006-01-06 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Fatty-acid-binding protein from the flight muscle of Locusta migratoria: evolutionary variations in fatty acid binding.
Biochemistry, 45, 2006
|
|
2M3Z
 
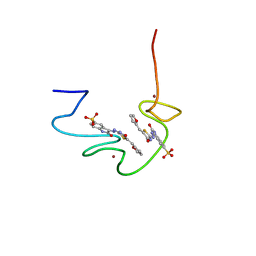 | |
2LVZ
 
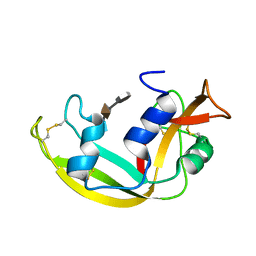 | | Solution structure of a Eosinophil Cationic Protein-trisaccharide heparin mimetic complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-propan-2-yl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Eosinophil cationic protein | | Authors: | Garcia Mayoral, M, Canales, A, Diaz, D, Lopez Prados, J, Moussaoui, M, de Paz, J, Angulo, J, Nieto, P, Jimenez Barbero, J, Boix, E, Bruix, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Insights into the glycosaminoglycan-mediated cytotoxic mechanism of eosinophil cationic protein revealed by NMR.
Acs Chem.Biol., 8, 2013
|
|
6Z50
 
 | | Crystal structure of CLK1 in complex with macrocycle ODS2003208 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-dimethyl-6-(oxan-4-yloxy)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CLK1 in complex with macrocycle ODS2003208
To Be Published
|
|
6YTE
 
 | | CLK1 bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
2M70
 
 | |
6YTW
 
 | | CLK3 bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
2LUP
 
 | |
6YTY
 
 | | CLK3 A319V mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
2LUQ
 
 | |
2FDY
 
 | | Microsomal P450 2A6 with the inhibitor Adrithiol bound | | Descriptor: | 4,4'-DIPYRIDYL DISULFIDE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
