2B4M
 
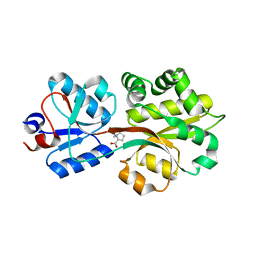 | | Crystal structure of the binding protein OpuAC in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding protein | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
5T7J
 
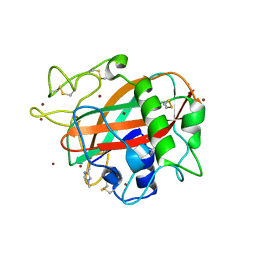 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3OFJ
 
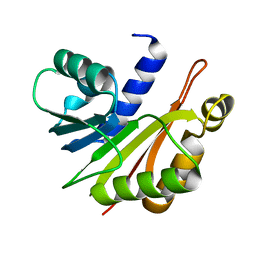 | | Crystal structure of N-methyltransferase NodS from Bradyrhizobium japonicum WM9 | | Descriptor: | Nodulation protein S | | Authors: | Cakici, O, Sikorski, M, Stepkowski, T, Bujacz, G, Jaskolski, M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of NodS N-Methyltransferase from Bradyrhizobium japonicum in Ligand-Free Form and as SAH Complex.
J.Mol.Biol., 404, 2010
|
|
4O6T
 
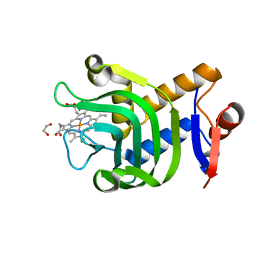 | | 1.25A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, pH 5.4) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
1M67
 
 | | Crystal Structure of Leishmania mexicana GPDH Complexed with Inhibitor 2-bromo-6-hydroxy-purine | | Descriptor: | 2-BROMO-6-HYDROXY-PURINE, Glycerol-3-phosphate dehydrogenase, PALMITIC ACID | | Authors: | Choe, J, Suresh, S, Wisedchaisri, G, Kennedy, K.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2002-07-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to
glycerol-3-phosphate dehydrogenase
Chem.Biol., 9, 2002
|
|
3N7A
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 2 | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N8K
 
 | | Type II dehydroquinase from Mycobacterium tuberculosis complexed with citrazinic acid | | Descriptor: | 2,6-dioxo-1,2,3,6-tetrahydropyridine-4-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3ND5
 
 | |
4G1V
 
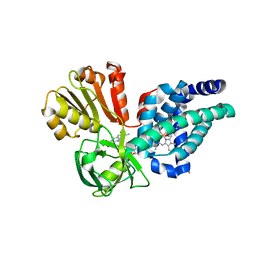 | | X-ray structure of yeast flavohemoglobin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoglobin, NITRITE ION, ... | | Authors: | El Hammi, E, Warkentin, E, Demmer, U, Baciou, L, Ermler, U. | | Deposit date: | 2012-07-11 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Active site analysis of yeast flavohemoglobin based on its structure with a small ligand or econazole.
Febs J., 279, 2012
|
|
4O6U
 
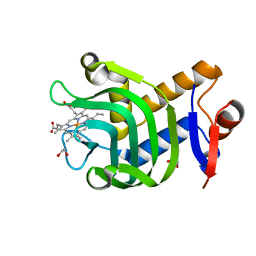 | | 0.89A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
3ND6
 
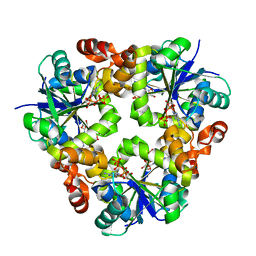 | |
1JDJ
 
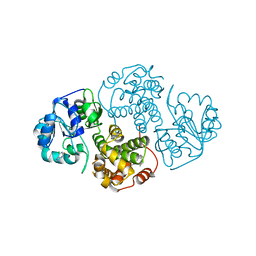 | | CRYSTAL STRUCTURE OF LEISHMANIA MEXICANA GLYCEROL-3-PHOSPHATE DEHYDROGENASE IN COMPLEX WITH 2-FLUORO-6-CHLOROPURINE | | Descriptor: | 6-CHLORO-2-FLUOROPURINE, GLYCEROL-3-PHOSPHATE DEHYDROGENASE, PENTADECANE | | Authors: | Suresh, S, Wisedchaisri, G, Kennedy, K.J, Verlinde, C.L.M.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2001-06-14 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to glycerol-3-phosphate dehydrogenase.
Chem.Biol., 9, 2002
|
|
3PN2
 
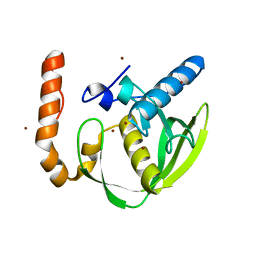 | |
4J9T
 
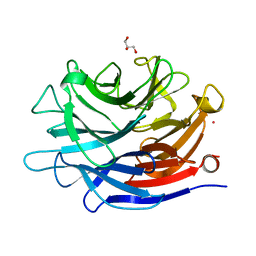 | | Crystal structure of a putative, de novo designed unnatural amino acid dependent metalloprotein, northeast structural genomics consortium target OR61 | | Descriptor: | ARSENIC, GLYCEROL, designed unnatural amino acid dependent metalloprotein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Mills, J.H, Khare, S.D, Everett, J.K, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
4JBL
 
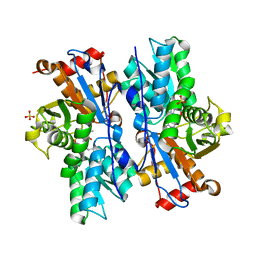 | |
4HBN
 
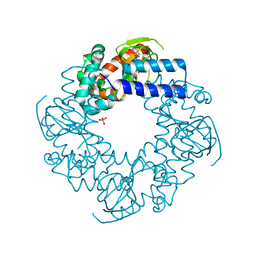 | | Crystal structure of the human HCN4 channel C-terminus carrying the S672R mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Marni, F, Wu, X, Su, Z, Musayev, F, Shrestha, S, Xie, C, Gao, W, Liu, Q, Zhou, L. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Local and Global Interpretations of a Disease-Causing Mutation near the Ligand Entry Path in Hyperpolarization-Activated cAMP-Gated Channel.
Structure, 20, 2012
|
|
4EMO
 
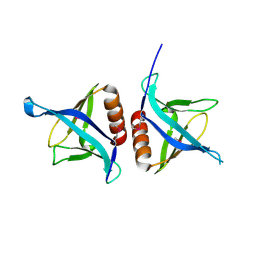 | | Crystal structure of the PH domain of SHARPIN | | Descriptor: | Sharpin | | Authors: | Stieglitz, B, Haire, L.F, Dikic, I, Rittinger, K. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of SHARPIN, a Subunit of a Large Multi-protein E3 Ubiquitin Ligase, Reveals a Novel Dimerization Function for the Pleckstrin Homology Superfold.
J.Biol.Chem., 287, 2012
|
|
3RO9
 
 | |
1SBR
 
 | | The structure and function of B. subtilis YkoF gene product: the complex with thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, CALCIUM ION, ykoF | | Authors: | Devedjiev, Y, Surendranath, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure and Ligand Binding Properties of the B.subtilis YkoF Gene Product, a Member of a Novel Family of Thiamin/HMP-binding Proteins
J.Mol.Biol., 343, 2004
|
|
1IUB
 
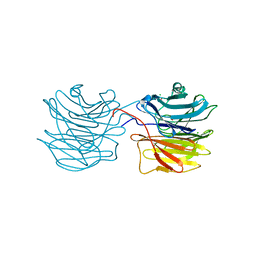 | | Fucose-specific lectin from Aleuria aurantia (Hg-derivative form) | | Descriptor: | CHLORIDE ION, Fucose-specific lectin, MERCURY (II) ION, ... | | Authors: | Fujihashi, M, Peapus, D.H, Kamiya, N, Nagata, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-01 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Fucose-Specific Lectin from Aleuria aurantia Binding Ligands at Three of Its Five Sugar Recognition Sites
Biochemistry, 42, 2003
|
|
2CUP
 
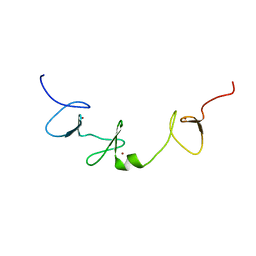 | | Solution structure of the Skeletal muscle LIM-protein 1 | | Descriptor: | Skeletal muscle LIM-protein 1, ZINC ION | | Authors: | Niraula, T.N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-27 | | Release date: | 2005-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Skeletal muscle LIM-protein 1
To be Published
|
|
2CPI
 
 | | Solution structure of the RNA recognition motif of CNOT4 | | Descriptor: | CCR4-NOT transcription complex subunit 4 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA recognition motif of CNOT4
To be Published
|
|
2CUC
 
 | | Solution structure of the SH3 domain of the mouse hypothetical protein SH3RF2 | | Descriptor: | SH3 domain containing ring finger 2 | | Authors: | Ohnishi, S, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the mouse hypothetical protein SH3RF2
To be Published
|
|
3CFN
 
 | |
2KF6
 
 | |
