1VSL
 
 | | ASV INTEGRASE CORE DOMAIN D64N MUTATION WITH ZINC CATION | | Descriptor: | PROTEIN (INTEGRASE), ZINC ION | | Authors: | Lubkowski, J, Yang, F, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inactivating mutations and pH-dependent activity of avian sarcoma virus integrase.
J.Biol.Chem., 273, 1998
|
|
1BVW
 
 | | CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Varrot, A, Davies, G.J, Schulein, M. | | Deposit date: | 1998-09-19 | | Release date: | 1999-09-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of the catalytic core domain of the family 6 cellobiohydrolase II, Cel6A, from Humicola insolens, at 1.92 A resolution.
Biochem.J., 337, 1999
|
|
2CMK
 
 | | CYTIDINE MONOPHOSPHATE KINASE IN COMPLEX WITH CYTIDINE-DI-PHOSPHATE | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PROTEIN (CYTIDINE MONOPHOSPHATE KINASE), SULFATE ION | | Authors: | Golinelli-Pimpaneau, B, Briozzo, P. | | Deposit date: | 1998-09-19 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Structure, 6, 1998
|
|
1BVY
 
 | | COMPLEX OF THE HEME AND FMN-BINDING DOMAINS OF THE CYTOCHROME P450(BM-3) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, PROTEIN (CYTOCHROME P450 BM-3), ... | | Authors: | Sevrioukova, I.F, Li, H, Zhang, H, Peterson, J.A, Poulos, T.L. | | Deposit date: | 1998-09-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3NCM
 
 | | NEURAL CELL ADHESION MOLECULE, MODULE 2, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (NEURAL CELL ADHESION MOLECULE, LARGE ISOFORM) | | Authors: | Jensen, P.H, Soroka, V, Thomsen, N.K, Berezin, V, Bock, E, Poulsen, F.M. | | Deposit date: | 1998-09-21 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of NCAM modules 1 and 2, basic elements in neural cell adhesion
Nat.Struct.Biol., 6, 1999
|
|
1BV2
 
 | | LIPID TRANSFER PROTEIN FROM RICE SEEDS, NMR, 14 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Poznanski, J, Sodano, P, Suh, S.W, Lee, J.Y, Ptak, M, Vovelle, F. | | Deposit date: | 1998-09-21 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a lipid transfer protein extracted from rice seeds. Comparison with homologous proteins.
Eur.J.Biochem., 259, 1999
|
|
427D
 
 | |
5KTQ
 
 | | LARGE FRAGMENT OF TAQ DNA POLYMERASE BOUND TO DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, PROTEIN (DNA POLYMERASE I) | | Authors: | Li, Y, Kong, Y, Korolev, S, Waksman, G. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates.
Protein Sci., 7, 1998
|
|
1BV4
 
 | | APO-MANNOSE-BINDING PROTEIN-C | | Descriptor: | PROTEIN (MANNOSE-BINDING PROTEIN-C) | | Authors: | Ng, K.K.-S, Weis, W.I. | | Deposit date: | 1998-09-22 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ca2+-dependent structural changes in C-type mannose-binding proteins.
Biochemistry, 37, 1998
|
|
1BWB
 
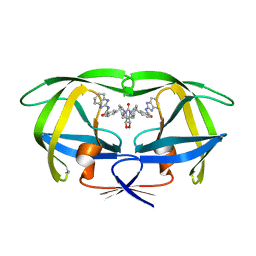 | | HIV-1 PROTEASE (V82F/I84V) DOUBLE MUTANT COMPLEXED WITH SD146 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6ALPHA,7ALPHA)]-3,3'-{{TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-DIYL]BIS(METHYLENE)]BIS[N-1H-BENZIMIDAZOL-2-YLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BV7
 
 | | COUNTERACTING HIV-1 PROTEASE DRUG RESISTANCE: STRUCTURAL ANALYSIS OF MUTANT PROTEASES COMPLEXED WITH XV638 AND SD146, CYCLIC UREA AMIDES WITH BROAD SPECIFICITIES | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1SXZ
 
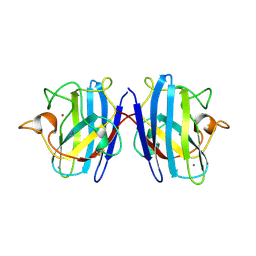 | | Reduced bovine superoxide dismutase at pH 5.0 complexed with azide | | Descriptor: | AZIDE ION, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W.R, Bruni, B, Orioli, P, Wilson, K.S, Mangani, S. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic determination of reduced bovine superoxide dismutase at pH 5.0 and of anion binding to its active site
J.Biol.Inorg.Chem., 3, 1998
|
|
1BWA
 
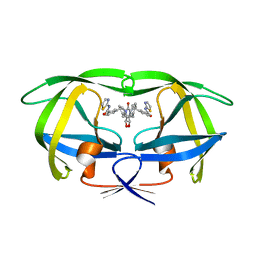 | | HIV-1 PROTEASE (V82F/I84V) DOUBLE MUTANT COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BWG
 
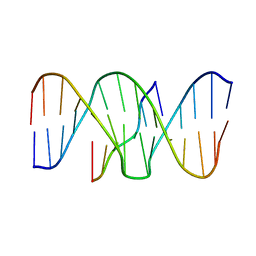 | | DNA TRIPLEX WITH 5' AND 3' JUNCTIONS, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*T)-3'), DNA (5'-D(*GP*AP*CP*TP*GP*AP*GP*AP*GP*AP*CP*GP*TP*A)-3'), DNA (5'-D(*TP*AP*CP*GP*TP*CP*TP*CP*TP*CP*AP*GP*TP*C)-3') | | Authors: | Asensio, J.L, Brown, T, Lane, A.N. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a parallel DNA triple helix with 5' and 3' triplex-duplex junctions.
Structure Fold.Des., 7, 1999
|
|
1BV9
 
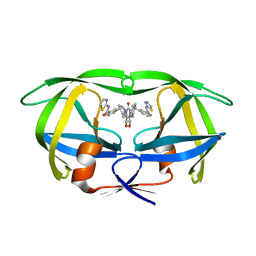 | | HIV-1 PROTEASE (I84V) COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BVZ
 
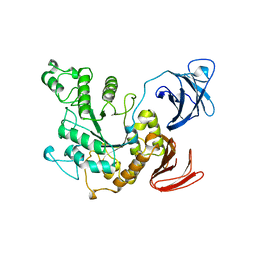 | | ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 | | Descriptor: | PROTEIN (ALPHA-AMYLASE II) | | Authors: | Kamitori, S, Kondo, S, Okuyama, K, Yokota, T, Shimura, Y, Tonozuka, T, Sakano, Y. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVAII) hydrolyzing cyclodextrins and pullulan at 2.6 A resolution.
J.Mol.Biol., 287, 1999
|
|
1BV8
 
 | |
1BV3
 
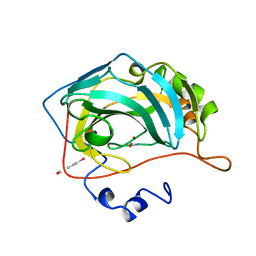 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH UREA | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, PROTEIN (CARBONIC ANHYDRASE II), UREA, ... | | Authors: | Briganti, F, Mangani, S, Scozzafava, A, Vernaglione, G, Supuran, C.T. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Carbonic anhydrase catalyzes cyanamide hydration to urea: is it mimicking the physiological reaction?
J.Biol.Inorg.Chem., 4, 1999
|
|
1SML
 
 | | METALLO BETA LACTAMASE L1 FROM STENOTROPHOMONAS MALTOPHILIA | | Descriptor: | PROTEIN (PENICILLINASE), ZINC ION | | Authors: | Ullah, J.H, Walsh, T.R, Taylor, I.A, Emery, D.C, Verma, C.S, Gamblin, S.J, Spencer, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution.
J.Mol.Biol., 284, 1998
|
|
1T1D
 
 | |
2FAP
 
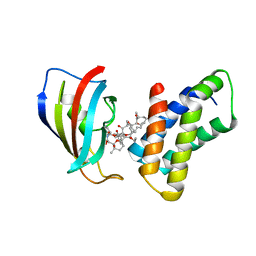 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-(C16)-ETHOXY RAPAMYCIN COMPLEX INTERACTING WITH HUMA | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FRAP | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2TIO
 
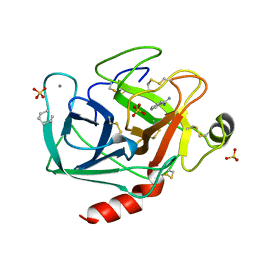 | | LOW PACKING DENSITY FORM OF BOVINE BETA-TRYPSIN IN CYCLOHEXANE | | Descriptor: | BENZAMIDINE, CALCIUM ION, HEXANE, ... | | Authors: | Huang, Q, Zhu, G, Tang, Q. | | Deposit date: | 1998-09-23 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray studies on two forms of bovine beta-trypsin crystals in neat cyclohexane.
Biochim.Biophys.Acta, 1429, 1998
|
|
3NSE
 
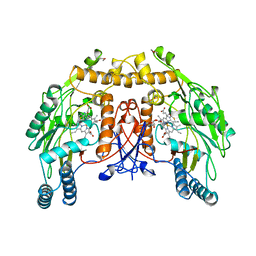 | | BOVINE ENOS, H4B-FREE, SEITU COMPLEX | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1998-09-23 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Cell(Cambridge,Mass.), 95, 1998
|
|
1BWC
 
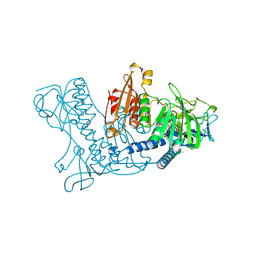 | | STRUCTURE OF HUMAN GLUTATHIONE REDUCTASE COMPLEXED with AJOENE INHIBITOR AND SUBVERSIVE SUBSTRATE | | Descriptor: | 3-(PROP-2-ENE-1-SULFINYL)-PROPENE-1-THIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gallwitz, H, Bonse, S, Martinez-Cruz, A, Schlichting, I, Schumacher, K, Krauth-Siegel, R.L. | | Deposit date: | 1998-09-23 | | Release date: | 1999-07-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ajoene is an inhibitor and subversive substrate of human glutathione reductase and Trypanosoma cruzi trypanothione reductase: crystallographic, kinetic, and spectroscopic studies.
J.Med.Chem., 42, 1999
|
|
1BWM
 
 | | A SINGLE-CHAIN T CELL RECEPTOR | | Descriptor: | PROTEIN (ALPHA-BETA T CELL RECEPTOR (TCR) (D10)) | | Authors: | Hare, B.J, Wyss, D.F, Reinherz, E.L, Wagner, G. | | Deposit date: | 1998-09-23 | | Release date: | 1999-07-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor.
Nat.Struct.Biol., 6, 1999
|
|
