2D57
 
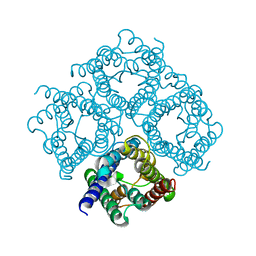 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
2D8X
 
 | |
1SOA
 
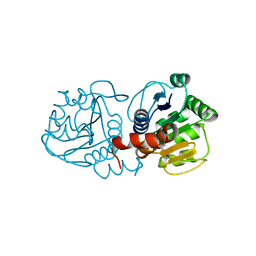 | | Human DJ-1 with sulfinic acid | | Descriptor: | RNA-binding protein regulatory subunit; oncogene DJ1 | | Authors: | Canet-Aviles, R, Wilson, M.A, Miller, D.W, Ahmad, R, McLendon, C, Bandyopadhyay, S, Baptista, M.J, Ringe, D, Petsko, G.A, Cookson, M.R. | | Deposit date: | 2004-03-13 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VFL
 
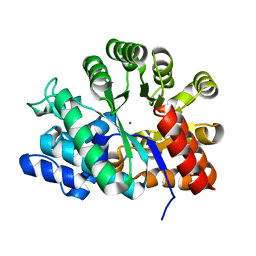 | | Adenosine deaminase | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2004-04-16 | | Release date: | 2005-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Compound Recognition by Adenosine Deaminase
Biochemistry, 44, 2005
|
|
1W7L
 
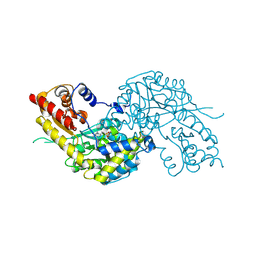 | | Crystal structure of human kynurenine aminotransferase I | | Descriptor: | KYNURENINE--OXOGLUTARATE TRANSAMINASE I, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
2EW5
 
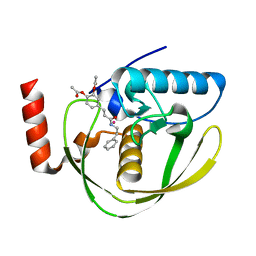 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | 4-{(1E)-3-OXO-3-[(2-PHENYLETHYL)AMINO]PROP-1-EN-1-YL}-1,2-PHENYLENE DIACETATE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2F0Y
 
 | |
1VEB
 
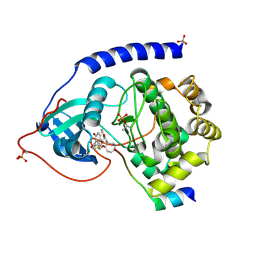 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 5 | | Descriptor: | (3R,4R)-N-{4-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZYLOXY]-AZEPAN-3-YL}-ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
2FFY
 
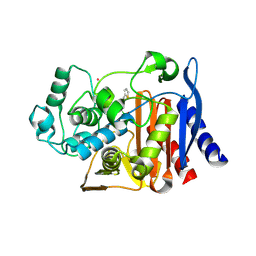 | | AmpC beta-lactamase N289A mutant in complex with a boronic acid deacylation transition state analog compound SM3 | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Chen, Y, Minasov, G, Roth, T.A, Prati, F, Shoichet, B.K. | | Deposit date: | 2005-12-20 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution
J.Am.Chem.Soc., 128, 2006
|
|
2F1S
 
 | | Crystal Structure of a Viral FLIP MC159 | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Li, F.-Y, Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2005-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Viral FLIP: INSIGHTS INTO FLIP-MEDIATED INHIBITION OF DEATH RECEPTOR SIGNALING.
J.Biol.Chem., 281, 2006
|
|
2FJP
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with an inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-{(1S,2S)-2-AMINO-1-[(DIMETHYLAMINO)CARBONYL]-3-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-3-OXOPROPYL}PHENYL)-1H-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-4-IUM, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W. | | Deposit date: | 2006-01-03 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (2S,3S)-3-Amino-4-(3,3-difluoropyrrolidin-1-yl)-N,N-dimethyl-4-oxo-2-(4-[1,2,4]triazolo[1,5-a]- pyridin-6-ylphenyl)butanamide: a selective alpha-amino amide dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
J.Med.Chem., 49, 2006
|
|
2F3J
 
 | |
2F7Z
 
 | | Protein Kinase A bound to (R)-1-(1H-Indol-3-ylmethyl)-2-(2-pyridin-4-yl-[1,7]naphtyridin-5-yloxy)-ehylamine | | Descriptor: | (1S)-1-(1H-INDOL-3-YLMETHYL)-2-(2-PYRIDIN-4-YL-[1,7]NAPHTYRIDIN-5-YLOXY)-EHYLAMINE, PKI, inhibitory peptide, ... | | Authors: | Li, Q, Woods, K.W, Thomas, S, Zhu, G.D, Packard, G, Fisher, J, Li, T, Gong, J, Dinges, J, Song, X, Abrams, J, Luo, Y, Johnson, E.F, Shi, Y, Liu, X, Klinghofer, V, Des Jong, R, Oltersdorf, T, Stoll, V.S, Jakob, C.G, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2005-12-01 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationship of 3,4'-bispyridinylethylenes: discovery of a potent 3-isoquinolinylpyridine inhibitor of protein kinase B (PKB/Akt) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8ZJV
 
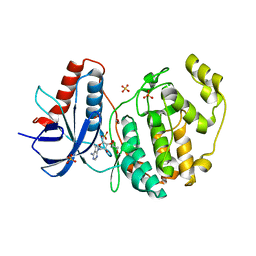 | | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Yang, Y, Fu, L, Chen, R, Cheng, H, Sun, X. | | Deposit date: | 2024-05-15 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin
To Be Published
|
|
2QME
 
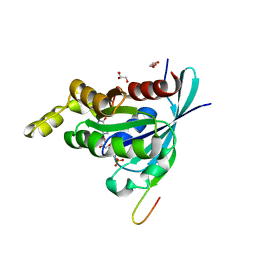 | | Crystal structure of human RAC3 in complex with CRIB domain of human p21-activated kinase 1 (PAK1) | | Descriptor: | CRIB domain of the Serine/threonine-protein kinase PAK 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J.M, Burgess-Brown, N, Bunkoczi, G, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 1 (PAK1).
To be Published
|
|
8W0T
 
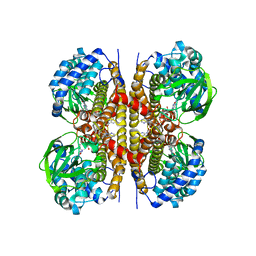 | | Human LCAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Long-chain specific acyl-CoA dehydrogenase, mitochondrial | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for expanded substrate specificities of human long chain acyl-CoA dehydrogenase and related acyl-CoA dehydrogenases.
Sci Rep, 14, 2024
|
|
8XT2
 
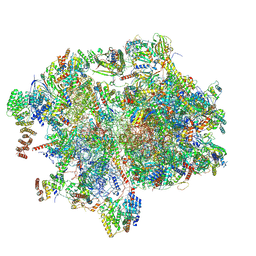 | | Cryo-EM structure of the human 55S mitoribosome with 10uM Tigecycline | | Descriptor: | 12s rRNA, 16s rRNA, 39S ribosomal protein L22, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8BRE
 
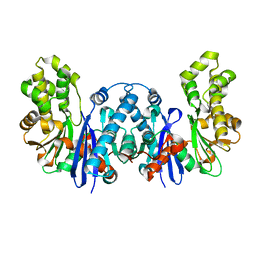 | |
9BRC
 
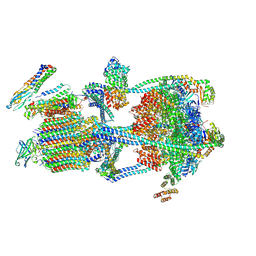 | |
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9RNT
 
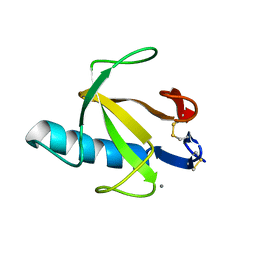 | |
8W9A
 
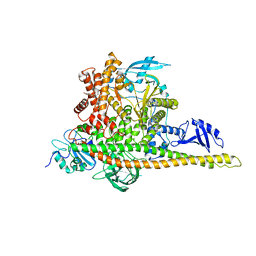 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site | | Descriptor: | 6-chloranyl-3-[[(1R)-1-[2-(1,3-dihydropyrrolo[3,4-c]pyridin-2-yl)-3,6-dimethyl-4-oxidanylidene-quinazolin-8-yl]ethyl]amino]pyridine-2-carboxylic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
8W6K
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BL03HB: Laue crystallography beamline at SSRF
To Be Published
|
|
8WLS
 
 | |
8W0Z
 
 | | Human LCAD complexed with Lauric Acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LAURIC ACID, Long-chain specific acyl-CoA dehydrogenase, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for expanded substrate specificities of human long chain acyl-CoA dehydrogenase and related acyl-CoA dehydrogenases.
Sci Rep, 14, 2024
|
|
