1GMM
 
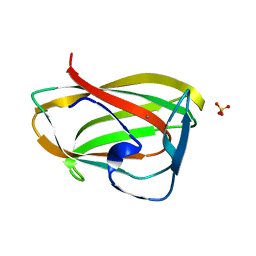 | | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | | Descriptor: | CALCIUM ION, CBM6, SODIUM ION, ... | | Authors: | Czjzek, M, Mosbah, A, Bolam, D, Allouch, J, Zamboni, V, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2001-09-19 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved.
J.Biol.Chem., 276, 2001
|
|
8P60
 
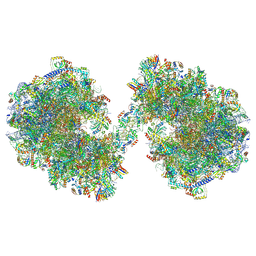 | | Spraguea lophii ribosome dimer | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2023-05-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (14.3 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
8P5D
 
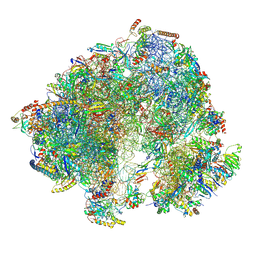 | | Spraguea lophii ribosome in the closed conformation by cryo sub tomogram averaging | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2023-05-23 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
3QPG
 
 | |
3QJU
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
1GOT
 
 | | HETEROTRIMERIC COMPLEX OF A GT-ALPHA/GI-ALPHA CHIMERA AND THE GT-BETA-GAMMA SUBUNITS | | Descriptor: | GT-ALPHA/GI-ALPHA CHIMERA, GT-BETA, GT-GAMMA, ... | | Authors: | Lambright, D.G, Sondek, J, Bohm, A, Skiba, N.P, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1996-08-07 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of a heterotrimeric G protein.
Nature, 379, 1996
|
|
3QN6
 
 | |
1GNE
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE OF SCHISTOSOMA JAPONICUM FUSED WITH A CONSERVED NEUTRALIZING EPITOPE ON GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Lim, K, Ho, J.X, Keeling, K, Gilliland, G.L, Ji, X, Ruker, F, Carter, D.C. | | Deposit date: | 1994-06-16 | | Release date: | 1994-11-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV.
Protein Sci., 3, 1994
|
|
3QRF
 
 | | Structure of a domain-swapped FOXP3 dimer | | Descriptor: | Forkhead box protein P3, MAGNESIUM ION, Nuclear factor of activated T-cells, ... | | Authors: | Bandukwala, H.S, Wu, Y, Feurer, M, Chen, Y, Barbosa, B, Ghosh, S, Stroud, J.C, Benoist, C, Mathis, D, Rao, A, Chen, L. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Domain-Swapped FOXP3 Dimer on DNA and Its Function in Regulatory T Cells.
Immunity, 34, 2011
|
|
3Q7J
 
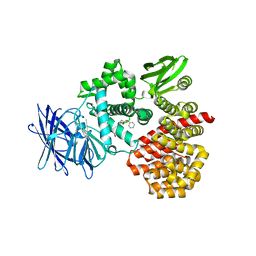 | | Engineered Thermoplasma Acidophilum F3 factor mimics human aminopeptidase N (APN) as a target for anticancer drug development | | Descriptor: | L-phenylalanyl-N6-[(benzyloxy)carbonyl]-N1-hydroxy-L-lysinamide, Tricorn protease-interacting factor F3, ZINC ION | | Authors: | Su, J, Wang, Q, Feng, J, Zhang, C, Zhu, D, We, T, Xu, W, Gu, L. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineered Thermoplasma acidophilum factor F3 mimics human aminopeptidase N (APN) as a target for anticancer drug development
Bioorg.Med.Chem., 19, 2011
|
|
3QAZ
 
 | | IL-2 mutant D10 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, Interleukin-2, ... | | Authors: | Levin, A.M, Bates, D.L, Ring, A.M, Lin, J.T, Su, L, Krieg, C, Bowman, G.R, Novick, P, Pande, V.S, Khort, H.E, Boyman, O, Gathman, C.G, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Exploiting a natural conformational switch to engineer an interleukin-2 'superkine'
Nature, 484, 2012
|
|
3QNF
 
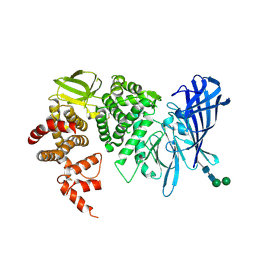 | | Crystal structure of the open state of human endoplasmic reticulum aminopeptidase 1 ERAP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, ZINC ION, ... | | Authors: | Vollmar, M, Kochan, G, Krojer, T, Harvey, D, Chaikuad, A, Allerston, C, Muniz, J.R.C, Raynor, J, Ugochukwu, E, Berridge, G, Wordsworth, B.P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QRA
 
 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
1JOL
 
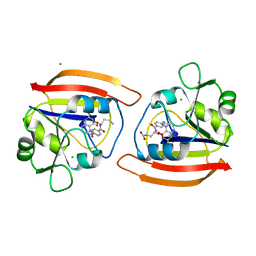 | | THE CRYSTAL STRUCTURE OF THE BINARY COMPLEX BETWEEN FOLINIC ACID (LEUCOVORIN) AND E. COLI DIHYDROFOLATE REDUCTASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Lee, H, Reyes, V.M, Kraut, J. | | Deposit date: | 1996-02-25 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of Escherichia coli dihydrofolate reductase complexed with 5-formyltetrahydrofolate (folinic acid) in two space groups: evidence for enolization of pteridine O4.
Biochemistry, 35, 1996
|
|
1JOM
 
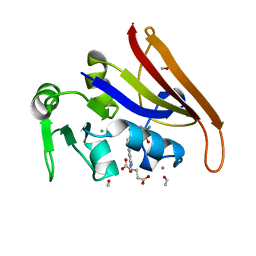 | | THE CRYSTAL STRUCTURE OF THE BINARY COMPLEX BETWEEN FOLINIC ACID (LEUCOVORIN) AND E. COLI DIHYDROFOLATE REDUCTASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Reyes, V.M, Lee, H, Kraut, J. | | Deposit date: | 1996-02-25 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Escherichia coli dihydrofolate reductase complexed with 5-formyltetrahydrofolate (folinic acid) in two space groups: evidence for enolization of pteridine O4.
Biochemistry, 35, 1996
|
|
3RQP
 
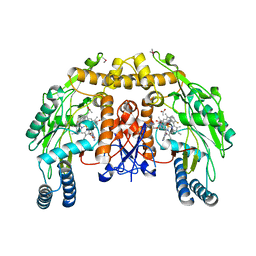 | | Structure of the endothelial nitric oxide synthase heme domain in complex with 6-{[(3R,4R)-4-(2-{[(2R/2S)-1-(3-fluorophenyl)propan-2-yl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(2R)-1-(3-fluorophenyl)propan-2-yl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2011-04-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cyclopropyl- and methyl-containing inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 21, 2013
|
|
3R0H
 
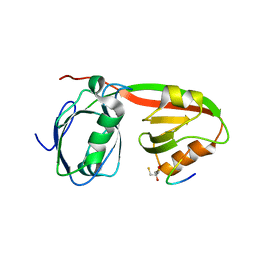 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
3RQK
 
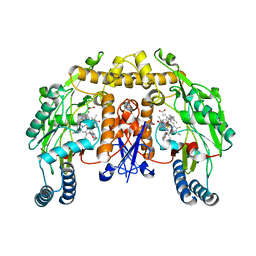 | | Structure of the neuronal nitric oxide synthase heme domain in complex with 4-methyl-6-{[(3R,4R)-4-(2-{[(1R,2S)-2-(3-methylphenyl)cyclopropyl]amino}ethoxy)pyrrolidin-3-yl]methyl}pyridin-2-amine and its isomer | | Descriptor: | 4-methyl-6-{[(3R,4R)-4-(2-{[(1R,2S)-2-(3-methylphenyl)cyclopropyl]amino}ethoxy)pyrrolidin-3-yl]methyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2011-04-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Cyclopropyl- and methyl-containing inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 21, 2013
|
|
3R0G
 
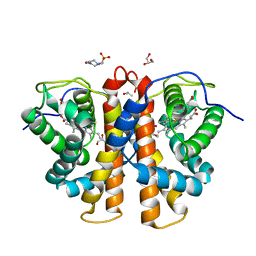 | | 3D Structure of Ferric Methanosarcina Acetivorans Protoglobin I149F mutant in Aquomet form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
1ITK
 
 | | Crystal structure of catalase-peroxidase from Haloarcula marismortui | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Yamada, Y, Fujiwara, T, Sato, T, Igarashi, N, Tanaka, N. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of catalase-peroxidase from Haloarcula marismortui.
Nat.Struct.Biol., 9, 2002
|
|
3RQL
 
 | | Structure of the neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((1S,2R/1R,2S)-2-(3-Clorophenyl)cyclopropylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(1R,2S)-2-(3-chlorophenyl)cyclopropyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2011-04-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Cyclopropyl- and methyl-containing inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 21, 2013
|
|
1E5B
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-07-24 | | Release date: | 2001-05-25 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
3OGD
 
 | | AlkA Undamaged DNA Complex: Interrogation of a G*:C base pair | | Descriptor: | 5'-D(*CP*AP*(BRU)P*GP*AP*CP*(BRU)P*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*CP*AP*TP*G)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
3OH9
 
 | | AlkA Undamaged DNA Complex: Interrogation of a T:A base pair | | Descriptor: | 5'-D(*AP*CP*AP*(BRU)P*GP*AP*AP*(BRU)P*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*TP*TP*CP*AP*TP*GP*T)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
1K8C
 
 | | Crystal structure of dimeric xylose reductase in complex with NADP(H) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
