4ZLF
 
 | | Cellobionic acid phosphorylase - cellobionic acid complex | | Descriptor: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
2ZON
 
 | | Crystal structure of electron transfer complex of nitrite reductase with cytochrome c | | Descriptor: | COPPER (II) ION, Dissimilatory copper-containing nitrite reductase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nojiri, M, Koteishi, H, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of inter-protein electron transfer for nitrite reduction in denitrification
Nature, 462, 2009
|
|
4Z13
 
 | | Recombinantly expressed latent aurone synthase (polyphenol oxidase) co-crystallized with hexatungstotellurate(VI) and soaked in H2O2 | | Descriptor: | 6-tungstotellurate(VI), Aurone synthase, COPPER (II) ION, ... | | Authors: | Molitor, C, Mauracher, S.G, Rompel, A. | | Deposit date: | 2015-03-26 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3LSW
 
 | | Aniracetam bound to the ligand binding domain of GluA3 | | Descriptor: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, GLUTAMIC ACID, GluA2 S1S2 domain, ... | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2010-02-13 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
3HN4
 
 | |
1MLB
 
 | | MONOCLONAL ANTIBODY FAB D44.1 RAISED AGAINST CHICKEN EGG-WHITE LYSOZYME | | Descriptor: | IGG1-KAPPA D44.1 FAB (HEAVY CHAIN), IGG1-KAPPA D44.1 FAB (LIGHT CHAIN) | | Authors: | Braden, B.C, Souchon, H, Eisele, J.-L, Bentley, G.A, Bhat, T.N, Navaza, J, Poljak, R.J. | | Deposit date: | 1995-03-08 | | Release date: | 1995-06-03 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of the free and the antigen-complexed Fab from monoclonal anti-lysozyme antibody D44.1.
J.Mol.Biol., 243, 1994
|
|
8EA8
 
 | | NKG2D complexed with inhibitor 4a | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5O2U
 
 | | Llama VHH in complex with p24 | | Descriptor: | Capsid protein p24, VHH 59H10 | | Authors: | Caillat, C, Verrips, T, Weissenhorn, W. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Unravelling the Molecular Basis of High Affinity Nanobodies against HIV p24: In Vitro Functional, Structural, and in Silico Insights.
ACS Infect Dis, 3, 2017
|
|
4ZLI
 
 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
8EA7
 
 | | NKG2D complexed with inhibitor 3g | | Descriptor: | (4M)-N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4-(1-methyl-1H-pyrazol-5-yl)-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, DI(HYDROXYETHYL)ETHER, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA5
 
 | | NKG2D complexed with inhibitor 1a | | Descriptor: | (3S,5aS,8aR)-3-benzyl-6-[(3,5-dichlorophenyl)methyl]-1,4-dimethyloctahydropyrrolo[3,2-e][1,4]diazepine-2,5-dione, NKG2-D type II integral membrane protein, TRIETHYLENE GLYCOL | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA9
 
 | | NKG2D complexed with inhibitor 4d | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-1-[3,5-bis(trifluoromethyl)phenyl]-2-(dimethylamino)-2-oxoethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAB
 
 | | NKG2D complexed with inhibitor 4f | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-2-oxo-1-[3-(trifluoromethyl)phenyl]-2-({4-[4-(trifluoromethyl)phenyl]pyridin-3-yl}amino)ethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA6
 
 | | NKG2D complexed with inhibitor 3e | | Descriptor: | N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAA
 
 | | NKG2D complexed with inhibitor 4e | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-{(1S)-2-(dimethylamino)-1-[3-methyl-5-(trifluoromethyl)phenyl]-2-oxoethyl}-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5O7O
 
 | | The crystal structure of DfoC, the desferrioxamine biosynthetic pathway acetyltransferase/Non-Ribosomal Peptide Synthetase (NRPS)-Independent Siderophore (NIS) from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | Desferrioxamine siderophore biosynthesis protein dfoC | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-09 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
4Z0Y
 
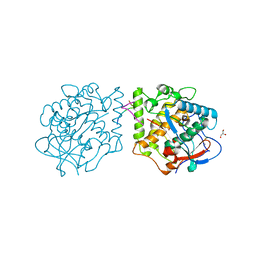 | | Active aurone synthase (polyphenol oxidase), copper B : sulfohistidine ~ 1.4 : 1 | | Descriptor: | Aurone synthase, COPPER (II) ION, GLYCEROL | | Authors: | Molitor, C, Mauracher, S.G, Rompel, A. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8DQ2
 
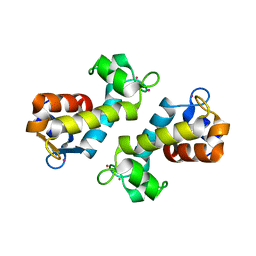 | | X-ray crystal structure of Hansschlegelia quercus lanmodulin (LanM) with lanthanum (III) bound at pH 7 | | Descriptor: | CITRIC ACID, EF-hand domain-containing protein, LANTHANUM (III) ION, ... | | Authors: | Jung, J.J, Lin, C.-Y, Boal, A.K. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhanced rare-earth separation with a metal-sensitive lanmodulin dimer.
Nature, 618, 2023
|
|
8DNG
 
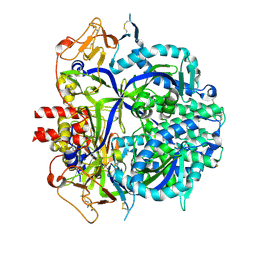 | |
8DNR
 
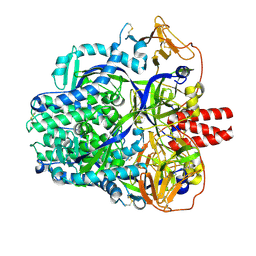 | |
4YK6
 
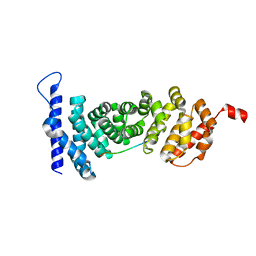 | | Crystal structure of APC-ARM in complexed with Amer1-A4 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
8DO4
 
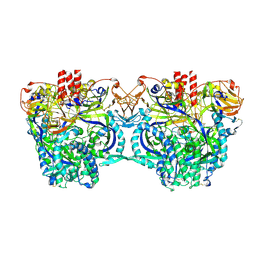 | |
4X1Z
 
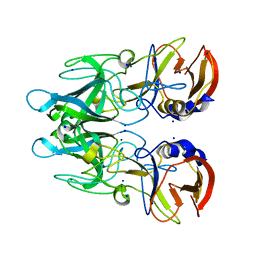 | | Crystal structure of RHDVb P domain in complex with H type 2 | | Descriptor: | SODIUM ION, VP1, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2014-11-25 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural analysis of a rabbit hemorrhagic disease virus binding to histo-blood group antigens.
J.Virol., 89, 2015
|
|
4Y5I
 
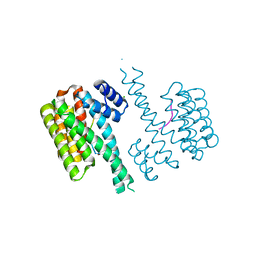 | | Crystal structure of C-terminal modified Tau peptide-hybrid 126B with 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Microtubule-associated protein tau | | Authors: | Leysen, S, Bartel, M, Milroy, L, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8EFI
 
 | | Helical reconstruction of the human cardiac actin-tropomyosin-myosin complex in the rigor form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Doran, M.H, Lehman, W, Rynkiewicz, M.J. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Myosin loop-4 is critical for optimal tropomyosin repositioning on actin during muscle activation and relaxation.
J.Gen.Physiol., 155, 2023
|
|
