6OVY
 
 | |
7XD8
 
 | | Crystal Structure of Dengue Virus Serotype 2 (DENV2) Polymerase Elongation Complex (Native Form) | | Descriptor: | GLYCEROL, NS5, RNA (30-mer), ... | | Authors: | Wu, J, Wang, X, Gong, P. | | Deposit date: | 2022-03-26 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XD9
 
 | | Crystal Structure of Dengue Virus serotype 2 (DENV2) Polymerase Elongation Complex (CTP Form) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wu, J, Wang, X, Gong, P. | | Deposit date: | 2022-03-26 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7KHI
 
 | | Escherichia coli RNA polymerase and rrnBP1 promoter complex with DksA/ppGpp | | Descriptor: | CHAPSO, DNA (28-MER), DNA (36-MER), ... | | Authors: | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2020-10-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
6OY6
 
 | |
7Q5B
 
 | | Cryo-EM structure of Ty3 retrotransposon targeting a TFIIIB-bound tRNA gene | | Descriptor: | DNA (19-MER), DNA (31-MER), DNA (34-MER), ... | | Authors: | Abascal-Palacios, G, Jochem, L, Pla-Prats, C, Beuron, F, Vannini, A. | | Deposit date: | 2021-11-03 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis of Ty3 retrotransposon integration at RNA Polymerase III-transcribed genes.
Nat Commun, 12, 2021
|
|
6OY7
 
 | |
6OVR
 
 | |
7DKH
 
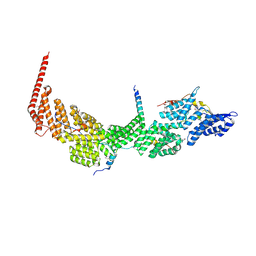 | | Crystal structure of the Ctr9/Paf1/Cdc73/Rtf1 quaternary complex | | Descriptor: | Cell division control protein 73, RNA polymerase II-associated protein 1, RNA polymerase-associated protein CTR9, ... | | Authors: | Chen, F.L, Liu, B.B, Guo, L, Li, D.F, Zhou, H, Long, J.F. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Core Module of the Yeast Paf1 Complex.
J.Mol.Biol., 434, 2021
|
|
6OMF
 
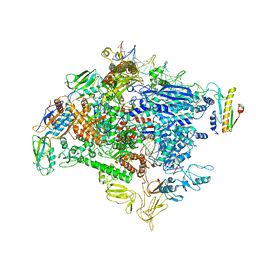 | | CryoEM structure of SigmaS-transcription initiation complex with activator Crl | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Jaramillo Cartagena, A, Darst, S.A, Campbell, E.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for transcription activation by Crl through tethering of sigmaSand RNA polymerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4WTJ
 
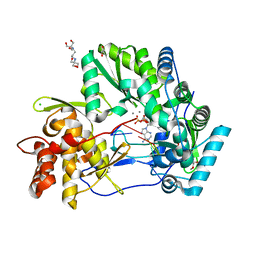 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AUCC, RNA PRIMER 5'-PGG, MN2+, AND ADP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
6OY5
 
 | |
4WTI
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-ACGG, RNA PRIMER 5'-PCC, MN2+, AND GDP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTK
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AGCC, RNA PRIMER 5'-PGG, MN2+, AND CDP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTL
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UACC, RNA PRIMER 5'-PGG, MN2+, AND UDP | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTM
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UAGG, RNA PRIMER 5'-PCC, MN2+, AND UDP | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, RNA PRIMER CC, ... | | Authors: | Edwards, T.E, Appleby, T.C, Fox III, D. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
6TYG
 
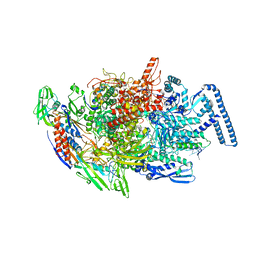 | | Crystal structure of MTB sigma L transcription initiation complex with 9 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5O7X
 
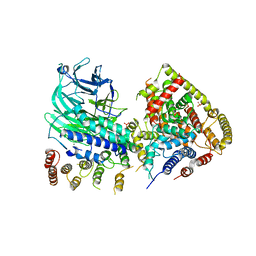 | | CRYSTAL STRUCTURE OF S. CEREVISIAE CORE FACTOR AT 3.2A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA polymerase I-specific transcription initiation factor RRN11, RNA polymerase I-specific transcription initiation factor RRN6, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-06-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
6TYF
 
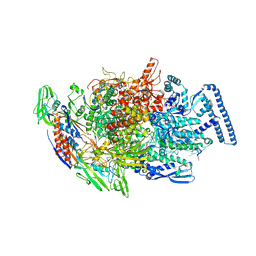 | | Crystal structure of MTB sigma L transcription initiation complex with 6 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4Y2C
 
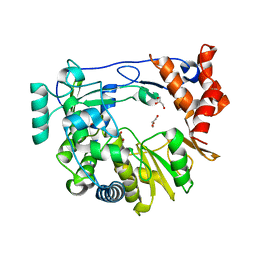 | | M300V 3D polymerase mutant of EMCV | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
6TYE
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 5 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PMJ
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
6PMI
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 1 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
7KHC
 
 | | Escherichia coli RNA polymerase and rrnBP1 promoter closed complex | | Descriptor: | CHAPSO, DNA (18 MER), DNA (63-MER), ... | | Authors: | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2020-10-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
7KHB
 
 | | Escherichia coli RNA polymerase and rrnBP1 promoter open complex | | Descriptor: | CHAPSO, DNA (60-MER), DNA (64-MER), ... | | Authors: | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2020-10-20 | | Release date: | 2020-10-28 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
