6IN7
 
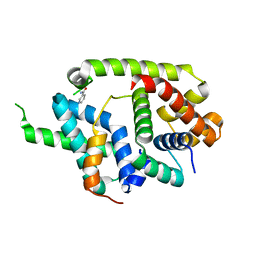 | | Crystal structure of AlgU in complex with MucA(cyto) | | Descriptor: | NICOTINAMIDE, RNA polymerase sigma-H factor, Sigma factor AlgU negative regulatory protein | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
5V0S
 
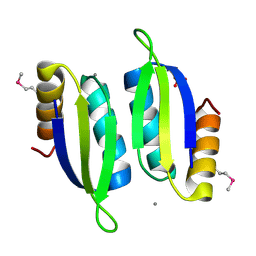 | | Crystal structure of the ACT domain of prephenate dehydrogenase tyrA from Bacillus anthracis | | Descriptor: | CALCIUM ION, Prephenate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Hou, J, Cymborowski, M.T, Otwinowski, Z, Kwon, K, Christendat, D, Gritsunov, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
6Q0N
 
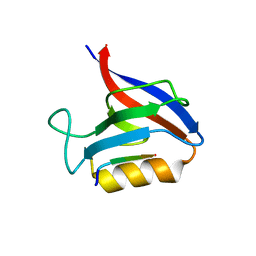 | | Structure of the Erbin PDB domain in complex with a high-affinity peptide | | Descriptor: | Erbin, peptide | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6TOR
 
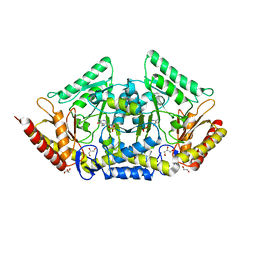 | | human O-phosphoethanolamine phospho-lyase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Ethanolamine-phosphate phospho-lyase, GLYCEROL | | Authors: | Vettraino, C, Donini, S, Parisini, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural characterization of human O-phosphoethanolamine phospho-lyase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6GIR
 
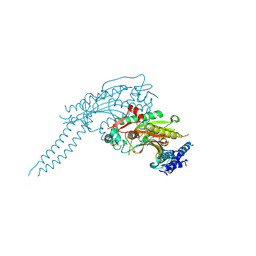 | | Arabidopsis thaliana cytosolic seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase, cytoplasmic | | Authors: | Kekez, I, Kekez, M, Rokov-Plavec, J, Matkovic-Calogovic, D. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Arabidopsis seryl-tRNA synthetase: the first crystal structure and novel protein interactor of plant aminoacyl-tRNA synthetase.
FEBS J., 286, 2019
|
|
6DCN
 
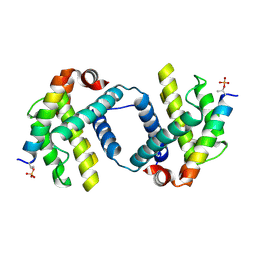 | | Bcl-xL complex with Beclin 1 BH3 domain T108pThr | | Descriptor: | BCL-xl protein, Beclin-1 | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6TQQ
 
 | |
7E3U
 
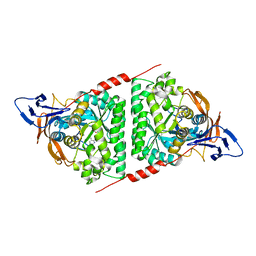 | | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-AU | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Yang, Y.C, Luo, R.H, Huang, Y.H, Huang, C.Y, Lin, E.S. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Molecular Insights into How the Dimetal Center in Dihydropyrimidinase Can Bind the Thymine Antagonist 5-Aminouracil: A Different Binding Mode from the Anticancer Drug 5-Fluorouracil.
Bioinorg Chem Appl, 2022, 2022
|
|
6GUJ
 
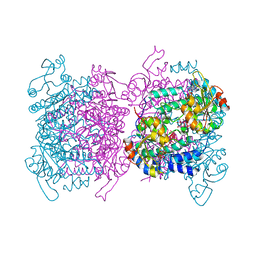 | | Molybdenum storage protein with two occupied ATP binding sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MOLYBDATE ION, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
7E50
 
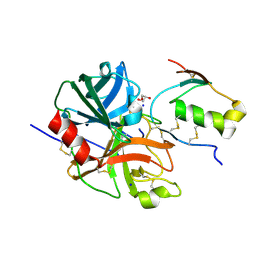 | | Crystal structure of human microplasmin in complex with kazal-type inhibitor AaTI | | Descriptor: | AAEL006007-PA, GLYCEROL, Plasminogen, ... | | Authors: | Varsha, A.W, Jobichen, C, Mok, Y.K. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Aedes aegypti trypsin inhibitor in complex with mu-plasmin reveals role for scaffold stability in Kazal-type serine protease inhibitor.
Protein Sci., 31, 2022
|
|
6DCO
 
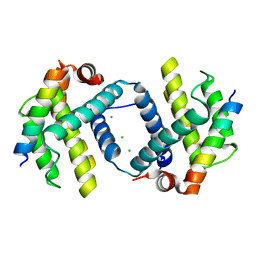 | | Bcl-xL complex with Beclin 1 BH3 domain T108D | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Beclin-1, CHLORIDE ION | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6A8M
 
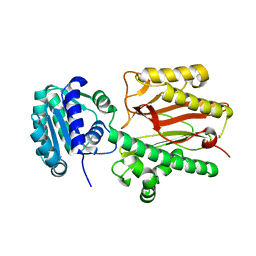 | | N-terminal domain of FACT complex subunit SPT16 from Eremothecium gossypii (Ashbya gossypii) | | Descriptor: | FACT complex subunit SPT16 | | Authors: | Gaur, N.K, Are, V.N, Durani, V, Ghosh, B, Kumar, A, Kulkarni, K, Makde, R.D. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolutionary conservation of protein dynamics: insights from all-atom molecular dynamics simulations of 'peptidase' domain of Spt16.
J.Biomol.Struct.Dyn., 2021
|
|
6GX4
 
 | | The molybdenum storage protein: with ATP/Mn2+ and with POM clusters formed under in vitro conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, MOLYBDATE ION, ... | | Authors: | Poppe, J, Bruenle, S, Hail, R, Ermler, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
9DIN
 
 | |
6GU5
 
 | | Mosto containing the core POM clusters | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MOLYBDATE ION, Molybdenum storage protein subunit alpha, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
6TQP
 
 | | Structural insight into tanapoxvirus mediated inhibition of apoptosis | | Descriptor: | 16L protein, Bcl-2-binding component 3, isoforms 1/2, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84940946 Å) | | Cite: | Structural insight into tanapoxvirus-mediated inhibition of apoptosis.
Febs J., 287, 2020
|
|
6H37
 
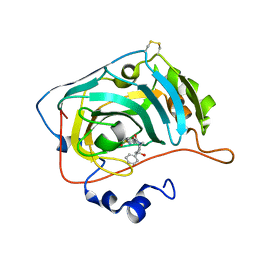 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenyl)-4-hydroxy-1-piperidine-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-(4-oxidanyl-4-phenyl-piperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6TRR
 
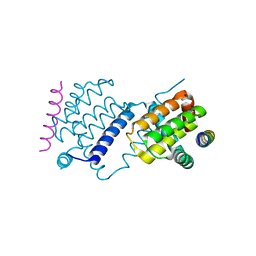 | |
6HDT
 
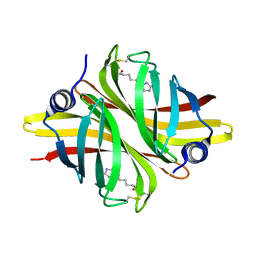 | |
7F25
 
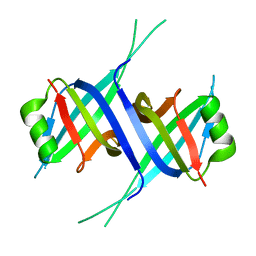 | |
6HDS
 
 | | Crystal Structure of apo short afifavidin | | Descriptor: | short afifavidin | | Authors: | Livnah, O, Avraham, O. | | Deposit date: | 2018-08-19 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of afifavidin reveals common features of molecular assemblage in the bacterial dimeric avidins.
FEBS J., 285, 2018
|
|
6HQI
 
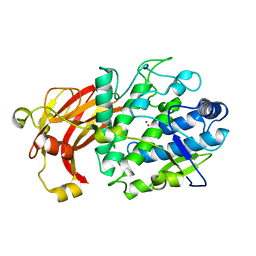 | | holo-form of polyphenol oxidase from Solanum lycopersicum | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Polyphenol oxidase A, ... | | Authors: | Kampatsikas, I, Bijelic, A, Rompel, A. | | Deposit date: | 2018-09-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of tomato polyphenol oxidases provide novel insights into their substrate specificity.
Sci Rep, 9, 2019
|
|
6HDV
 
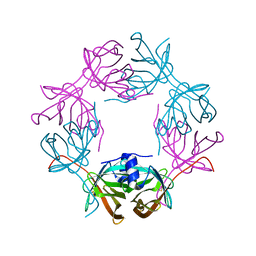 | |
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
6UE0
 
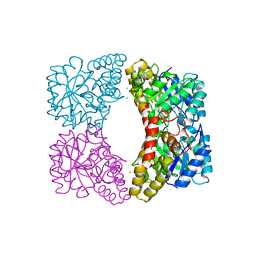 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
