5MRQ
 
 | | Arabidopsis thaliana IspD Asp262Ala Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5JGS
 
 | | Human carbonic anhydrase II (F131Y/L198A) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3QS7
 
 | |
5O6S
 
 | |
2HCH
 
 | |
5JDV
 
 | | Human carbonic anhydrase II (F131W) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2REG
 
 | | ABC-transporter choline binding protein in complex with choline | | Descriptor: | CHOLINE ION, PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Schimtt, L, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Bremer, E. | | Deposit date: | 2007-09-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
5JEH
 
 | | Human carbonic anhydrase II (L198A) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2YME
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
1IXQ
 
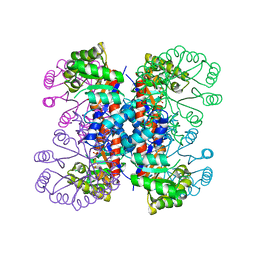 | | Enzyme-Phosphate2 Complex of Pyridoxine 5'-Phosphate synthase | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate Synthase | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
4P0I
 
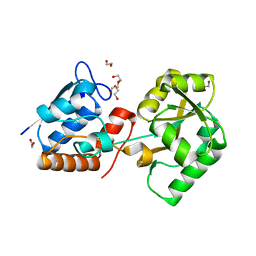 | | Structure of the PBP NocT | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Nopaline-binding periplasmic protein | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
4GX2
 
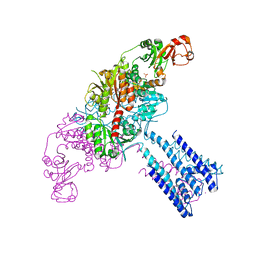 | | GsuK channel bound to NAD | | Descriptor: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-09-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
3MNO
 
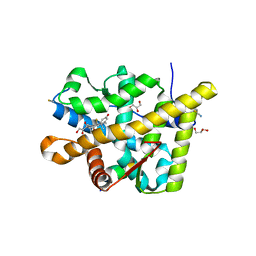 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, F608S) mutations at 1.55A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
1JBM
 
 | |
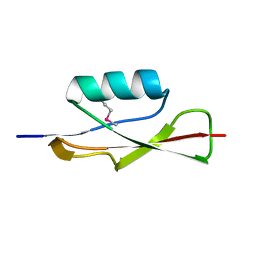
3OUV
 
 | |
1JF3
 
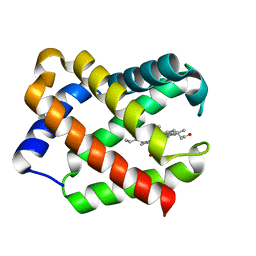 | | Crystal Structure Of Component III Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component III | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2001-06-20 | | Release date: | 2002-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer
Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
3GRU
 
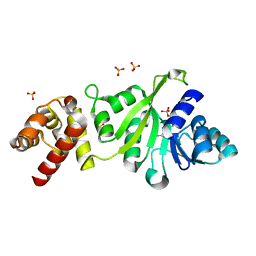 | |
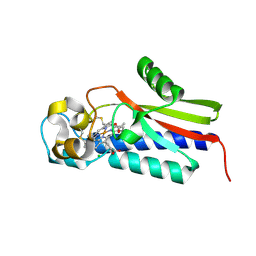
5B82
 
 | |
1JF4
 
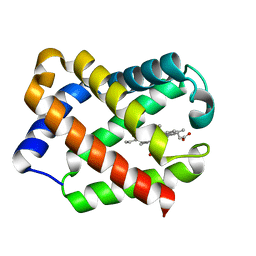 | | Crystal Structure Of Component IV Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component IV | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2001-06-20 | | Release date: | 2002-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer
Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
4GX1
 
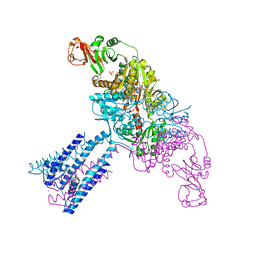 | | Crystal structure of the GsuK bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-09-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
3NY1
 
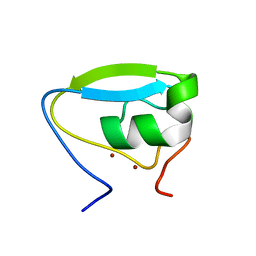 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1IXO
 
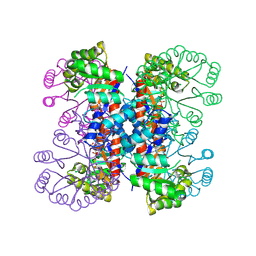 | | Enzyme-analogue substrate complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | Pyridoxine 5'-Phosphate synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
2RIN
 
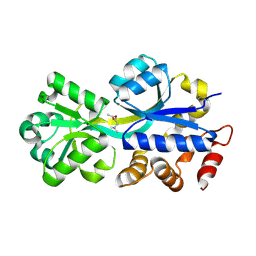 | | ABC-transporter choline binding protein in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
2CT2
 
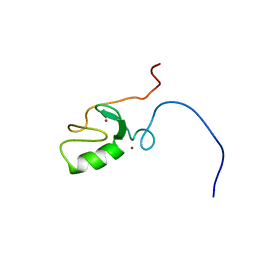 | | Solution Structure of the RING domain of the Tripartite motif protein 32 | | Descriptor: | Tripartite motif protein 32, ZINC ION | | Authors: | Miyamoto, K, Tochio, N, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RING domain of the Tripartite motif protein 32
To be Published
|
|
4GX0
 
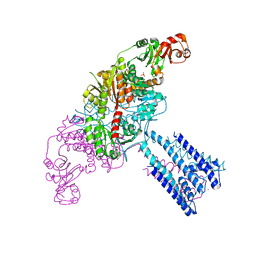 | | Crystal structure of the GsuK L97D mutant | | Descriptor: | CALCIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-09-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
