7DD0
 
 | |
7MBJ
 
 | |
4L9S
 
 | | Crystal Structure of H-Ras G12C, GDP-bound | | Descriptor: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
6F1O
 
 | | Orthorhombic Lysozyme crystallized at 298 K and pH 4.5 | | Descriptor: | CHLORIDE ION, Lysozyme C, PHOSPHATE ION | | Authors: | Camara-Artigas, A. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Orthorhombic lysozyme crystallization at acidic pH values driven by phosphate binding.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7MRI
 
 | | Crystal structure of N63T yeast iso-1-cytochrome c | | Descriptor: | Cytochrome c isoform 1, HEME C | | Authors: | Lei, H, Bowler, B.E, Evenson, G.E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Effect on intrinsic peroxidase activity of substituting coevolved residues from Omega-loop C of human cytochrome c into yeast iso-1-cytochrome c.
J.Inorg.Biochem., 232, 2022
|
|
7MRP
 
 | |
4LE0
 
 | | Crystal structure of the receiver domain of DesR in complex with beryllofluoride and magnesium | | Descriptor: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
6F5E
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, DD_D12_10_47, Mitogen-activated protein kinase 8 | | Authors: | Wu, Y, Mittl, P.R, Honegger, A, Batyuk, A, Plueckthun, A. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_10_47 in complex JNK1a1 and JIP1 peptide
To be published
|
|
6FD8
 
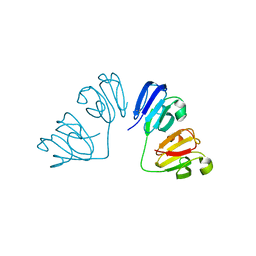 | | Gamma-s crystallin dimer | | Descriptor: | Beta-crystallin S | | Authors: | Mabbitt, P.D, Thorn, D.C, Jackson, C.J, Carver, J.A. | | Deposit date: | 2017-12-22 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure and Stability of the Disulfide-Linked gamma S-Crystallin Dimer Provide Insight into Oxidation Products Associated with Lens Cataract Formation.
J. Mol. Biol., 431, 2019
|
|
6F60
 
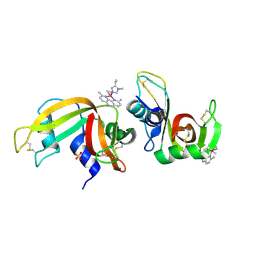 | | The x-ray structure of bovine pancreatic ribonuclease in complex with a five-coordinate platinum(II) compound containing a sugar ligand | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, five-coordinate platinum(II) compound | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2017-12-04 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Five-Coordinate Platinum(II) Compounds Containing Sugar Ligands: Synthesis, Characterization, Cytotoxic Activity, and Interaction with Biological Macromolecules.
Inorg Chem, 57, 2018
|
|
4LDJ
 
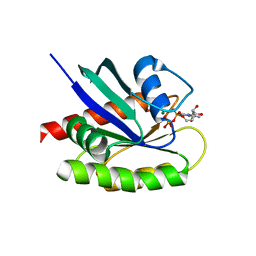 | | Crystal Structure of a GDP-bound G12C Oncogenic Mutant of Human GTPase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2013-06-24 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LDS
 
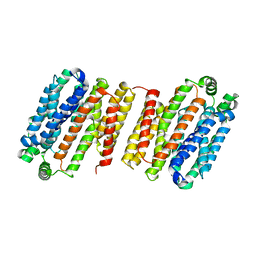 | |
4LE9
 
 | | Crystal structure of a chimeric c-Src-SH3 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, TRIETHYLENE GLYCOL | | Authors: | Camara-Artigas, A, Martinez-Rodriguez, S, Ortiz-Salmeron, E, Martin-Garcia, J.M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | 3D domain swapping in a chimeric c-Src SH3 domain takes place through two hinge loops.
J.Struct.Biol., 186, 2014
|
|
4LL9
 
 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
6F76
 
 | | Antibody derived (Abd-8) small molecule binding to KRAS. | | Descriptor: | 4-(2,3-dihydro-1,4-benzodioxin-5-yl)-~{N}-[3-[(dimethylamino)methyl]phenyl]-2-methoxy-aniline, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Bery, N, Cruz-Migoni, A, Quevedo, C.E, Phillips, S.V.E, Carr, S, Rabbitts, T.H. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions.
Elife, 7, 2018
|
|
7D7V
 
 | |
4LX8
 
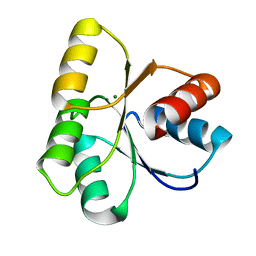 | |
7D0W
 
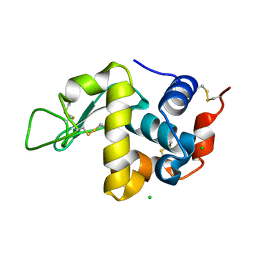 | |
7M1K
 
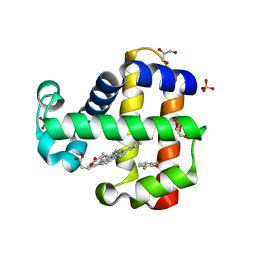 | | Crystal structure of dehaloperoxidase B in complex with 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2021-03-13 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Bridging the functional gap between reactivity and inhibition in dehaloperoxidase B from Amphitrite ornata: Mechanistic and structural studies with 2,4- and 2,6-dihalophenols.
J.Inorg.Biochem., 236, 2022
|
|
7M9W
 
 | | HIV-1 Protease (I84V) in Complex with NR02-73 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(1S)-1-hydroxy-2,3-dihydro-1H-indene-5-sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with NR02-73
To Be Published
|
|
7M9S
 
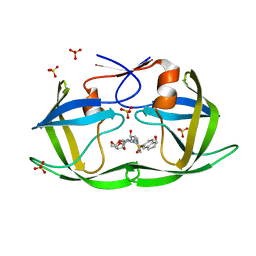 | | HIV-1 Protease WT (NL4-3) in Complex with NR01-141 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(1S)-1-hydroxy-2,3-dihydro-1H-indene-5-sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | HIV-1 Protease WT (NL4-3) in Complex with NR01-141
To Be Published
|
|
6FJB
 
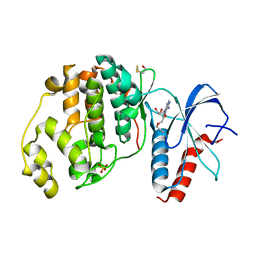 | |
7LXL
 
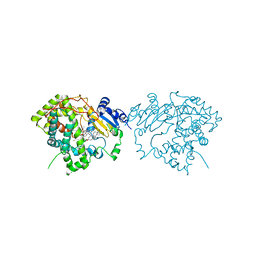 | | Crystal structure of human CYP3A4 bound to the testosterone dimer | | Descriptor: | 17alpha-hydroxy-7alpha-[(2Z)-4-(17beta-hydroxy-3-oxo-8alpha-androst-4-en-7beta-yl)but-2-en-1-yl]-8alpha,10alpha,13alpha,14beta-androst-4-en-3-one, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2021-03-04 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Innovative C 2 -symmetric testosterone and androstenedione dimers: Design, synthesis, biological evaluation on prostate cancer cell lines and binding study to recombinant CYP3A4.
Eur.J.Med.Chem., 220, 2021
|
|
4M1Y
 
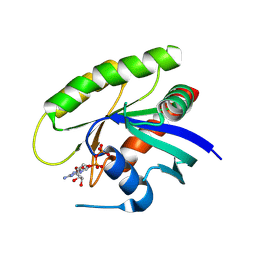 | | Crystal Structure of small molecule vinylsulfonamide 15 covalently bound to K-Ras G12C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, N-{1-[N-(5,7-dichloro-2,1,3-benzothiadiazol-4-yl)glycyl]piperidin-4-yl}ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
6ETO
 
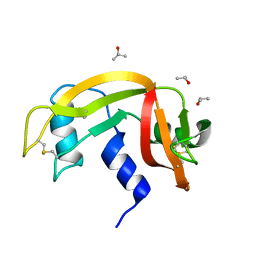 | | Atomic resolution structure of RNase A (data collection 5) | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Caterino, M, Vergara, A, Merlino, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 2018
|
|
