8BLR
 
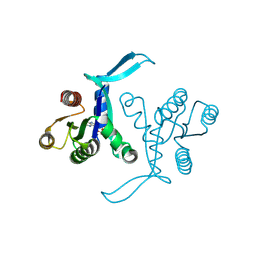 | | G13D mutant of KRAS4b (2-169) bound to GDP with the switch-I in fully open conformation | | Descriptor: | GTPase KRas, N-terminally processed, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Moche, M, Jungholm, O, Strandback, E, Ampah-Korsah, H, Nyman, T, Orwar, O. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel druggable space in human KRAS G13D discovered using structural bioinformatics and a P-loop targeting monoclonal antibody.
Sci Rep, 14, 2024
|
|
8CPR
 
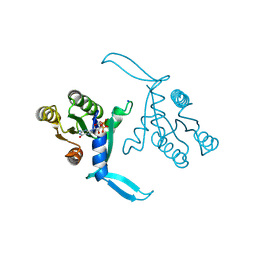 | | G13D mutant of KRAS4b (2-169) bound to GDP with the switch-I in fully open conformation crystallized in sodium potassium phosphate buffer | | Descriptor: | GTPase KRas, N-terminally processed, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Moche, M, Jungholm, O, Strandback, E, Ampah-Korsah, H, Nyman, T, Orwar, O. | | Deposit date: | 2023-03-03 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel druggable space in human KRAS G13D discovered using structural bioinformatics and a P-loop targeting monoclonal antibody.
Sci Rep, 14, 2024
|
|
1L6B
 
 | |
3QB7
 
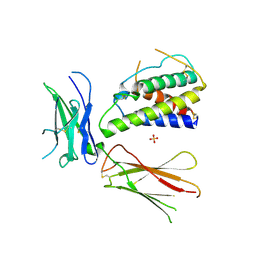 | | Interleukin-4 mutant RGA bound to cytokine receptor common gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Bates, D.L, Junttila, I.S, Creusot, R.J, Moraga, I, Lupardus, P, Fathman, C.G, Paul, W.E, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines.
Nat.Chem.Biol., 8, 2012
|
|
2G1E
 
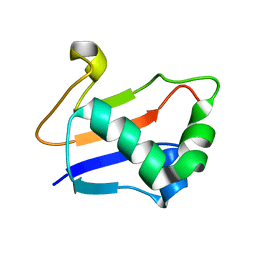 | | Solution Structure of TA0895 | | Descriptor: | hypothetical protein Ta0895 | | Authors: | Yeo, I.Y, Hong, E, Jung, J, Lee, W, Yee, A, Arrowsmith, C.H. | | Deposit date: | 2006-02-14 | | Release date: | 2006-12-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of TA0895, a MoaD homologue from Thermoplasma acidophilum
Proteins, 65, 2006
|
|
1BG7
 
 | | LOCALIZED UNFOLDING AT THE JUNCTION OF THREE FERRITIN SUBUNITS. A MECHANISM FOR IRON RELEASE? | | Descriptor: | CALCIUM ION, FERRITIN | | Authors: | Takagi, H, Shi, D, Ha, Y, Allewell, N.M, Theil, E.C. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Localized unfolding at the junction of three ferritin subunits. A mechanism for iron release?
J.Biol.Chem., 273, 1998
|
|
8JOY
 
 | | Plk1 polo-box domain bound to HPV4 L2 residues 251-257 with pThr255 | | Descriptor: | Peptide from Minor capsid protein L2, Serine/threonine-protein kinase PLK1 | | Authors: | Ku, B, Jung, S. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structures of Plk1 Polo-Box Domain Bound to the Human Papillomavirus Minor Capsid Protein L2-Derived Peptide.
J.Microbiol, 61, 2023
|
|
8JOQ
 
 | |
3IGT
 
 | |
1JUC
 
 | | Crystal Structure Analysis of a Holliday Junction Formed by CCGGTACCGG | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3' | | Authors: | Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Cardin, C.J. | | Deposit date: | 2001-08-24 | | Release date: | 2002-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of a new crystal form of the four-way Holliday junction formed by the DNA sequence d(CCGGTACCGG)2: sequence versus lattice?
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1R5S
 
 | | Connexin 43 Carboxyl Terminal Domain | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
1RK8
 
 | | Structure of the cytosolic protein PYM bound to the Mago-Y14 core of the exon junction complex | | Descriptor: | CALCIUM ION, CG8781-PA protein, Mago nashi protein, ... | | Authors: | Bono, F, Ebert, J, Guettler, T, Izaurralde, E, Conti, E. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the interaction of PYM with
the Mago-Y14 core of the exon junction complex
EMBO Rep., 5, 2004
|
|
4QOY
 
 | |
1KRQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CAMPYLOBACTER JEJUNI FERRITIN | | Descriptor: | ferritin | | Authors: | Hortolan, L, Saintout, N, Granier, G, Langlois d'Estaintot, B, Manigand, C, Mizunoe, Y, Wai, S.N, Gallois, B, Precigoux, G. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURE OF CAMPYLOBACTER JEJUNI FERRITIN AT 2.7 A RESOLUTION
To be Published
|
|
3IRQ
 
 | | Crystal structure of a Z-Z junction | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IRR
 
 | | Crystal Structure of a Z-Z junction (with HEPES intercalating) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*A*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*G*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
398D
 
 | | 3'-DNA-RNA-5' JUNCTION FORMED DURING INITIATION OF MINUS-STRAND SYNTHESIS OF HIV REPLICATION | | Descriptor: | DNA/RNA (5'-R(*GP*CP*CP*AP)-D(*CP*TP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*UP*GP*GP*C)-3') | | Authors: | Mueller, U, Meier, G, Mochi-Onori, A, Cellai, L, Heumann, H. | | Deposit date: | 1998-05-04 | | Release date: | 1998-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of an eight-base pair duplex containing the 3'-DNA-RNA-5' junction formed during initiation of minus-strand synthesis of HIV replication.
Biochemistry, 37, 1998
|
|
2NPO
 
 | | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Acetyltransferase | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3LER
 
 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-15 | | Release date: | 2010-01-26 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
2HYI
 
 | | Structure of the human exon junction complex with a trapped DEAD-box helicase bound to RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*U)-3', MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Andersen, C.B.F, Le Hir, H, Andersen, G.R. | | Deposit date: | 2006-08-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA.
Science, 313, 2006
|
|
3IZ1
 
 | | C-alpha model fitted into the EM structure of Cx26M34A | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3IZ2
 
 | | C-alpha model fitted into the EM structure of Cx26M34Adel2-7 | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
2J0Q
 
 | | The crystal structure of the Exon Junction Complex at 3.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Maintains a Stable Grip on Mrna.
Cell(Cambridge,Mass.), 126, 2006
|
|
2J0S
 
 | | The crystal structure of the Exon Junction Complex at 2.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP *UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
467D
 
 | |
