1WWD
 
 | | NMR structure determined for MLV NC complex with RNA sequence AACAGU | | Descriptor: | 5'-R(P*AP*AP*CP*AP*GP*U)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
1WWE
 
 | | NMR Structure Determined for MLV NC complex with RNA Sequence UUUUGCU | | Descriptor: | 5'-R(P*UP*UP*UP*UP*GP*CP*U)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
4E8B
 
 | |
2J7U
 
 | | Dengue virus NS5 RNA dependent RNA polymerase domain | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
7BPG
 
 | | Structure of serinol nucleic acid - RNA complex | | Descriptor: | CALCIUM ION, RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3'), SNA (S-(F7R)(F7X)(F7O)(F7R)(F7X)(F7O)(F7R)(F7U)-R) | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
7BPF
 
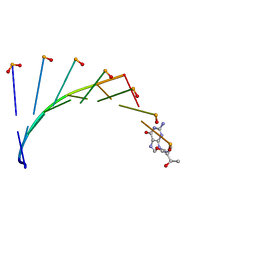 | | Structure of L-threoninol nucleic acid - RNA complex | | Descriptor: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
1GO3
 
 | | Structure of an archeal homolog of the eukaryotic RNA polymerase II RPB4/RPB7 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE SUBUNIT E, DNA-DIRECTED RNA POLYMERASE SUBUNIT F | | Authors: | Todone, F, Brick, P, Werner, F, Weinzierl, R.O.J, Onesti, S. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an Archaeal Homolog of the Eukaryotic RNA Polymerase II Rpb4/Rpb7 Complex
Mol.Cell, 8, 2001
|
|
2XFM
 
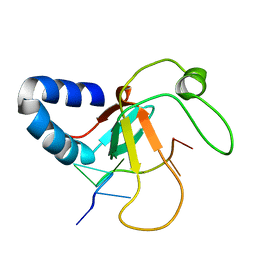 | | Complex structure of the MIWI Paz domain bound to methylated single stranded RNA | | Descriptor: | 5'-R(*AP*CP*CP*GP*AP*CP*UP*(OMU)P)-3', PIWI-LIKE PROTEIN 1 | | Authors: | Simon, B, Kirkpatrick, J.P, Eckhardt, S, Sehr, P, Andrade-Navarro, M.A, Pillai, R.S, Carlomagno, T. | | Deposit date: | 2010-05-27 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition of 2'-O-Methylated 3'-End of Pirna by the Paz Domain of a Piwi Protein.
Structure, 19, 2011
|
|
1ZCI
 
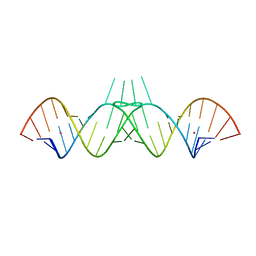 | | HIV-1 DIS RNA subtype F- monoclinic form | | Descriptor: | 5'-R(*CP*(5BU)P*UP*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', POTASSIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2005-04-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
2AB5
 
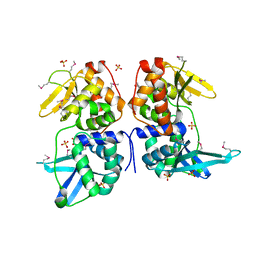 | | bI3 LAGLIDADG Maturase | | Descriptor: | SULFATE ION, mRNA maturase | | Authors: | Longo, A, Leonard, C.W, Bassi, G.S, Berndt, D, Krahn, J.M, Hall, T.M, Weeks, K.M. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase
Nat.Struct.Mol.Biol., 12, 2005
|
|
1TA0
 
 | | Three-dimensional structure of a RNA-polymerase II binding protein with associated ligand. | | Descriptor: | CITRIC ACID, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Kamenski, T, Heilmeier, S, Meinhart, T, Cramer, P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanism of RNA Polymerase II CTD Phosphatases.
Mol.Cell, 15, 2004
|
|
2L94
 
 | | Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA_(45-MER) | | Authors: | Marcheschi, R.J, Tonelli, M, Kumar, A, Butcher, S.E. | | Deposit date: | 2011-01-29 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 Frameshift Site RNA Bound to a Small Molecule Inhibitor of Viral Replication.
Acs Chem.Biol., 6, 2011
|
|
1XDN
 
 | | High resolution crystal structure of an editosome enzyme from trypanosoma brucei: RNA editing ligase 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA editing ligase MP52 | | Authors: | Deng, J, Schnaufer, A, Salavati, R, Stuart, K.D, Hol, W.G. | | Deposit date: | 2004-09-07 | | Release date: | 2004-12-07 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1.
J.Mol.Biol., 343, 2004
|
|
2W4R
 
 | | Crystal structure of the regulatory domain of human LGP2 | | Descriptor: | MERCURY (II) ION, PROBABLE ATP-DEPENDENT RNA HELICASE DHX58, SULFATE ION | | Authors: | Pippig, D.A, Hellmuth, J.C, Cui, S, Kirchhofer, A, Lammens, K, Lammens, A, Schmidt, A, Rothenfusser, S, Hopfner, K.P. | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Regulatory Domain of the Rig-I Family ATPase Lgp2 Senses Double-Stranded RNA.
Nucleic Acids Res., 37, 2009
|
|
1B2Z
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4BLW
 
 | | Crystal structure of Escherichia coli 23S rRNA (A2030-N6)- methyltransferase RlmJ in complex with S-adenosylhomocysteine (AdoHcy) and Adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Punekar, A.S, Liljeruhm, J, Shepherd, T.R, Forster, A.C, Selmer, M. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Insights Into the Molecular Mechanism of Rrna M6A Methyltransferase Rlmj.
Nucleic Acids Res., 41, 2013
|
|
1XEQ
 
 | | Crystal tructure of RNA binding domain of influenza B virus non-structural protein | | Descriptor: | BROMIDE ION, Nonstructural protein NS1 | | Authors: | Khan, J.A, Yin, C, Krug, R.M, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-11 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses.
J.Biol.Chem., 282, 2007
|
|
4BLV
 
 | | Crystal structure of Escherichia coli 23S rRNA (A2030-N6)- methyltransferase RlmJ in complex with S-adenosylmethionine (AdoMet) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Punekar, A.S, Liljeruhm, J, Shepherd, T.R, Forster, A.C, Selmer, M. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights Into the Molecular Mechanism of Rrna M6A Methyltransferase Rlmj.
Nucleic Acids Res., 41, 2013
|
|
1XP7
 
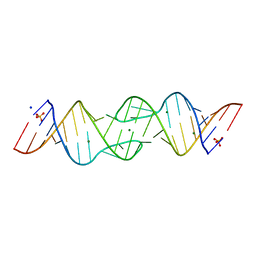 | | HIV-1 subtype F genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1XPF
 
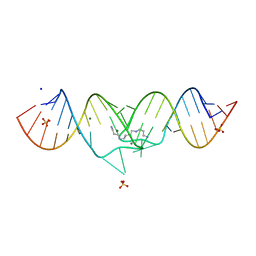 | | HIV-1 subtype A genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1B20
 
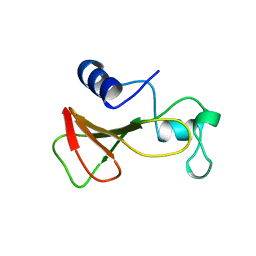 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1XPE
 
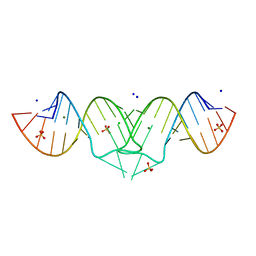 | | HIV-1 subtype B genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
3IUY
 
 | | Crystal structure of DDX53 DEAD-box domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX53 | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
1YY0
 
 | | Crystal structure of an RNA duplex containing a 2'-amine substitution and a 2'-amide product produced by in-crystal acylation at a C-A mismatch | | Descriptor: | 5'-R(*GP*CP*AP*GP*AP*(A5M)P*UP*UP*AP*AP*AP*UP*CP*UP*GP*C)-3', 5'-R(*GP*CP*AP*GP*AP*(M5M)P*UP*UP*AP*AP*AP*UP*CP*UP*GP*C)-3', CALCIUM ION | | Authors: | Gherghe, C.M, Krahn, J.M, Weeks, K.M. | | Deposit date: | 2005-02-23 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures, reactivity and inferred acylation transition States for 2'-amine substituted RNA.
J.Am.Chem.Soc., 127, 2005
|
|
1Y3O
 
 | | HIV-1 DIS RNA subtype F- Mn soaked | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION, SODIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-11-26 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
