8C43
 
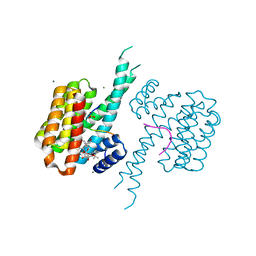 | | Ternary structure of 14-3-3sigma, strep-tagged PKA-responsive ERa phosphopeptide, and Fusicoccin-A. | | Descriptor: | 14-3-3 protein sigma, De-acetylated Fusicoccin, Estrogen Receptor alpha phosphopeptide, ... | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2022-12-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis and dual ligand regulation of tetrameric estrogen receptor alpha /14-3-3 zeta protein complex.
J.Biol.Chem., 299, 2023
|
|
1FCS
 
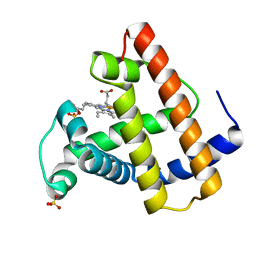 | | CRYSTAL STRUCTURE OF A DISTAL SITE DOUBLE MUTANT OF SPERM WHALE MYOGLOBIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Rizzi, M, Bolognesi, M, Coda, A, Cutruzzola, F, Travaglini Allocatelli, C, Brancaccio, A, Brunori, M. | | Deposit date: | 1993-04-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a distal site double mutant of sperm whale myoglobin at 1.6 A resolution.
FEBS Lett., 320, 1993
|
|
8C42
 
 | |
4N38
 
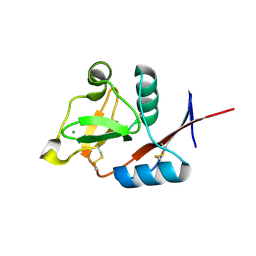 | | Structure of langerin CRD I313 D288 complexed with GlcNAc-beta1-3Gal-beta1-4GlcNAc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, C-type lectin domain family 4 member K, CALCIUM ION, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
8C3Z
 
 | |
3WFE
 
 | | Reduced and cyanide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CYANIDE ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
1KKQ
 
 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
4Z9J
 
 | | Apo-Tar from E. coli | | Descriptor: | Methyl-accepting chemotaxis protein II | | Authors: | Mise, T. | | Deposit date: | 2015-04-10 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Analysis of the Ligand-Binding Domain of the Aspartate Receptor Tar from Escherichia coli
Biochemistry, 55, 2016
|
|
4N36
 
 | | Structure of langerin CRD I313 D288 complexed with Me-GlcNAc | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
4N35
 
 | | Structure of langerin CRD I313 complexed with GlcNAc-beta1-3Gal-beta1-4Glc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, C-type lectin domain family 4 member K, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
7NVQ
 
 | | Aerosol-soaked human cdk2 crystals with Staurosporine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, STAUROSPORINE | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Aerosol-based ligand soaking of reservoir-free protein crystals.
J.Appl.Crystallogr., 54, 2021
|
|
4RXE
 
 | |
6O14
 
 | |
4RXC
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Homorisedronate BPH-6 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [1-hydroxy-3-(pyridin-3-yl)propane-1,1-diyl]bis(phosphonic acid) | | Authors: | Cao, R, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Farnesyl diphosphate synthase inhibitors with unique ligand-binding geometries.
ACS Med Chem Lett, 6, 2015
|
|
7YRT
 
 | |
3WFB
 
 | | Reduced cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
2GN2
 
 | |
3WFC
 
 | | Reduced and carbonmonoxide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
5PZO
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE C316N IN COMPLEX WITH 2-(4-FLUOROPHENYL)-N-METHYL-5-[3-({2-METHYL-1-OXO-1-[(1,3,4-THIADIAZOL-2-YL)AMINO]PROPAN-2-YL}CARBAMOYL)PHENYL]-1-BENZOFURAN-3-CARBOXAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-(4-fluorophenyl)-N-methyl-5-[3-({2-methyl-1-oxo-1-[(1,3,4-thiadiazol-2-yl)amino]propan-2-yl}carbamoyl)phenyl]-1-benzofuran-3-carboxamide, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
4FE7
 
 | | structure of xylose-binding transcription activator xylR | | Descriptor: | Xylose operon regulatory protein, alpha-D-xylopyranose | | Authors: | Ni, L, Schumacher, M.A. | | Deposit date: | 2012-05-29 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: an AraC DNA-binding family member with a LacI/GalR ligand-binding domain.
Nucleic Acids Res., 41, 2013
|
|
4FE4
 
 | | Crystal structure of apo E. coli XylR | | Descriptor: | Xylose operon regulatory protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2012-05-29 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: an AraC DNA-binding family member with a LacI/GalR ligand-binding domain.
Nucleic Acids Res., 41, 2013
|
|
3V56
 
 | | Re-refinement of PDB entry 1OSG - Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold - reveals an additonal copy of the peptide. | | Descriptor: | BR3 derived peptive, SULFATE ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Smart, O.S, Womack, T.O, Flensburg, C, Keller, P, Sharff, A, Paciorek, W, Vonrhein, C, Bricogne, G. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5YVA
 
 | | Structure of CaMKK2 in complex with CKI-010 | | Descriptor: | 3-(1H-tetrazol-5-yl)-10lambda~6~-thioxanthene-9,10,10-trione, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2020-06-10 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
2JGS
 
 | | Circular permutant of avidin | | Descriptor: | BIOTIN, CIRCULAR PERMUTANT OF AVIDIN | | Authors: | Maatta, J.A.E, Hytonen, V.P, Airenne, T.T, Niskanen, E, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-02-14 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Modification of Ligand-Binding Preference of Avidin by Circular Permutation and Mutagenesis.
Chembiochem, 9, 2008
|
|
5PZN
 
 | |
