2FD0
 
 | | HIV-1 DIS kissing-loop in complex with lividomycin | | Descriptor: | (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, CHLORIDE ION, HIV-1 DIS RNA, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
3WOF
 
 | | Crystal structure of P23-45 gp39 (6-132) bound to Thermus thermophilus RNA polymerase beta-flap domain | | Descriptor: | DNA-directed RNA polymerase subunit beta, Putative uncharacterized protein | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
3WOE
 
 | | Crystal structure of P23-45 gp39 (6-109) bound to Thermus thermophilus RNA polymerase beta-flap domain | | Descriptor: | DNA-directed RNA polymerase subunit beta, Putative uncharacterized protein | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
8BN6
 
 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL3021 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-morpholin-4-yl-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2022-11-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Dual Inhibitors of Bacterial Topoisomerases with Broad-Spectrum Antibacterial Activity and In Vivo Efficacy against Vancomycin-Intermediate Staphylococcus aureus .
J.Med.Chem., 66, 2023
|
|
3N4M
 
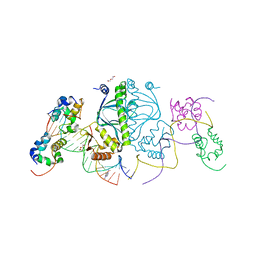 | | E. coli RNA polymerase alpha subunit C-terminal domain in complex with CAP and DNA | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lara-Gonzalez, S, Birktoft, J.J, Lawson, C.L. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.987 Å) | | Cite: | The RNA Polymerase alpha Subunit Recognizes the DNA Shape of the Upstream Promoter Element.
Biochemistry, 59, 2020
|
|
8BG1
 
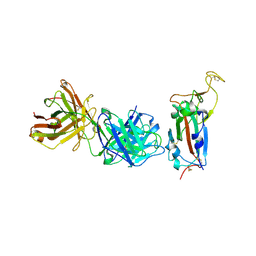 | | Crystal structure of the SARS-CoV-2 S RBD in complex with pT1511 scFV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hansen, G, Ssebyatika, G.L, Krey, T. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Activity of broadly neutralizing antibodies against sarbecoviruses: a trade-off between SARS-CoV-2 variants and distant coronaviruses?
To be published
|
|
8BEC
 
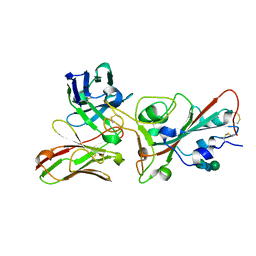 | | Crystal structure of the SARS-CoV-2 S RBD in complex with pT1375 scFV | | Descriptor: | SULFATE ION, Spike protein S1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hansen, G, Ssebyatika, G.L, Krey, T. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity of broadly neutralizing antibodies against sarbecoviruses: a trade-off between SARS-CoV-2 variants and distant coronaviruses?
To be published
|
|
8BG2
 
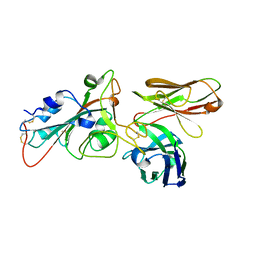 | |
3ADJ
 
 | |
4Y27
 
 | |
3O8B
 
 | |
1I4B
 
 | |
7K95
 
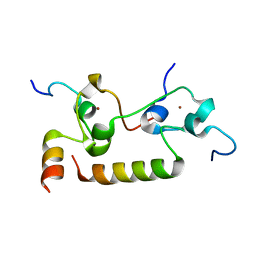 | | Crystal structure of human CPSF30 in complex with hFip1 | | Descriptor: | Isoform 2 of Cleavage and polyadenylation specificity factor subunit 4, Pre-mRNA 3'-end-processing factor FIP1, ZINC ION | | Authors: | Hamilton, K, Tong, L. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism for the interaction between human CPSF30 and hFip1.
Genes Dev., 34, 2020
|
|
6VZC
 
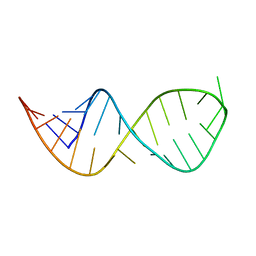 | |
1IDV
 
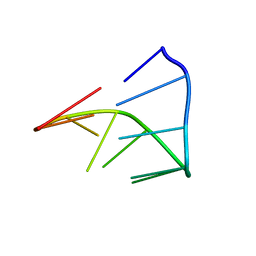 | |
3D6V
 
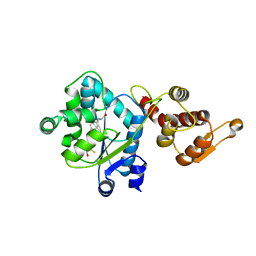 | | Crystal structure of 4-(trifluoromethyldiazirinyl)phenylalanyl-tRNA synthetase | | Descriptor: | 4-(2,2,2-TRIFLUOROETHYL)-L-PHENYLALANINE, BETA-MERCAPTOETHANOL, Tyrosyl-tRNA synthetase | | Authors: | Liu, W, Tippmann, E, Mack, A.V, Schultz, P.G. | | Deposit date: | 2008-05-20 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A genetically encoded diazirine photocrosslinker in Escherichia coli
ChemBioChem, 8, 2007
|
|
1M5K
 
 | |
4JVY
 
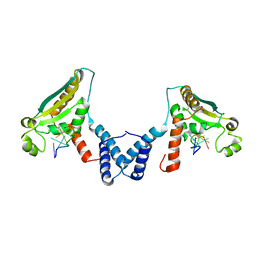 | | Structure of the STAR (signal transduction and activation of RNA) domain of GLD-1 bound to RNA | | Descriptor: | Female germline-specific tumor suppressor gld-1, RNA (5'-R(P*CP*UP*AP*AP*CP*AP*A)-3') | | Authors: | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-08 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
4JGN
 
 | |
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | Descriptor: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2M84
 
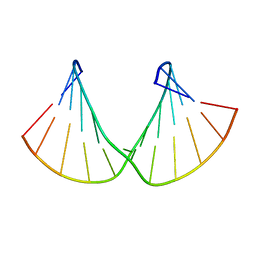 | | Structure of 2'F-RNA/2'F-ANA chimeric duplex | | Descriptor: | 2'F-RNA/2'F-ANA CHIMERIC DUPLEX | | Authors: | Martin-Pintado, N, Deleavey, G, Portella, G, Campos, R, Orozco, M, Damha, M, Gonzalez, C. | | Deposit date: | 2013-05-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone FCHO Hydrogen Bonds in 2'F-Substituted Nucleic Acids.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4CSG
 
 | |
6AIB
 
 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3F73
 
 | | Alignment of guide-target seed duplex within an argonaute silencing complex | | Descriptor: | ARGONAUTE, DNA (5'-D(P*DTP*DGP*DAP*DGP*DGP*DTP*DAP*DGP*DTP*DAP*DGP*DGP*DTP*DTP*DGP*DTP*DA*DTP*DAP*DGP*DT)-3'), MAGNESIUM ION, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Juranek, S, Tuschl, T, Patel, D.J. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex.
Nature, 456, 2008
|
|
6AIC
 
 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
