1AT1
 
 | |
7U3T
 
 | | [F223] Self-assembling tensegrity triangle with two turns, two turns and three turns of DNA per axis by extension with P1 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P, Zhu, E. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4.69 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U4R
 
 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-propyl-5,6-dihydrothieno[2,3-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 3,3-dimethyl-1-{[(9aM)-9-propyl-5,6-dihydrothieno[2,3-h]quinazolin-2-yl]sulfanyl}butan-2-one, Dual specificity protein phosphatase 10 | | Authors: | Gannam, Z.T.K, Jamali, H, Gentzel, E, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
8OLJ
 
 | | Crystal structure of Archaeoglobus fulgidus AfAgo-N protein representing N-L1-L2 domains | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, Archaeoglobus fulgidus AfAgo-N protein representing N-L1-L2 domains, ... | | Authors: | Manakova, E.N, Zaremba, M, Grazulis, S. | | Deposit date: | 2023-03-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
3KSB
 
 | | Detailed structural insight into the DNA cleavage complex of type IIA topoisomerases (re-sealed form) | | Descriptor: | 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*TP*GP*AP*CP*TP*AP*TP*GP*CP*AP*CP*GP*TP*AP*AP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*TP*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3', DNA topoisomerase 4 subunit A, ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-11-21 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
3KYL
 
 | |
1CZI
 
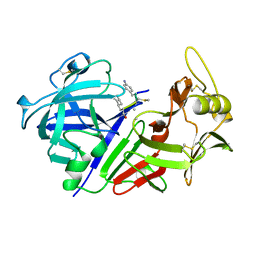 | | CHYMOSIN COMPLEX WITH THE INHIBITOR CP-113972 | | Descriptor: | CHYMOSIN, CP-113972 (NORSTATINE-S-METHYL CYSTEINE-IODO-PHENYLALANINE-PROLINE) | | Authors: | Groves, M.R, Dhanaraj, V, Pitts, J.E, Badasso, M, Hoover, D, Nugent, P, Blundell, T.L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 2.3 A resolution structure of chymosin complexed with a reduced bond inhibitor shows that the active site beta-hairpin flap is rearranged when compared with the native crystal structure.
Protein Eng., 11, 1998
|
|
4UWM
 
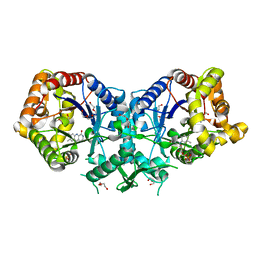 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6WPS
 
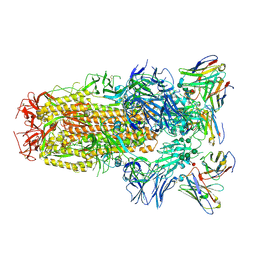 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
5KMG
 
 | | Near-atomic cryo-EM structure of PRC1 bound to the microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kellogg, E.H, Howes, S, Ti, S.-C, Ramirez-Aportela, E, Kapoor, T.M, Chacon, P, Nogales, E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic cryo-EM structure of PRC1 bound to the microtubule.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6O2X
 
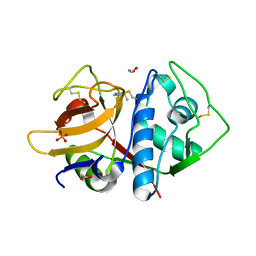 | | Structure of cruzain bound to MMTS inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | Authors: | Silva, E.B, Dall, E, Ferreira, R.S, Brandstetter, H. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6QTN
 
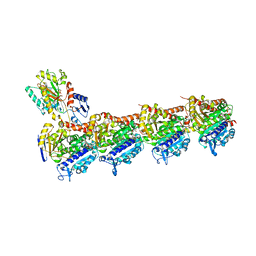 | | Tubulin-cyclostreptin complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Balaguer, F.d.A, Muehlethaler, T, Estevez-Gallego, J, Calvo, E, Gimenez-Abian, J.F, Risinger, A.L, Sorensen, E.J, Vanderwal, C.D, Altmann, K.-H, Mooberry, S.L, Steinmetz, M.O, Oliva, M.A, Prota, A.E, Diaz, J.F. | | Deposit date: | 2019-02-25 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Cyclostreptin-Tubulin Adduct: Implications for Tubulin Activation by Taxane-Site Ligands.
Int J Mol Sci, 20, 2019
|
|
8BYE
 
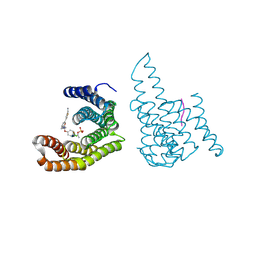 | |
7OMF
 
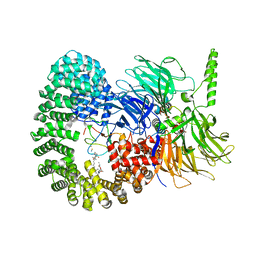 | | Structure of a minimal SF3B core in complex with sudemycin D6 (form I) | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
8BYB
 
 | |
8BWJ
 
 | |
8BWZ
 
 | |
8BX4
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1074392) | | Descriptor: | 14-3-3 protein sigma, 2-[3,4-bis(chloranyl)phenoxy]-~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]-2-methyl-propanamide, ERalpha peptide | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BYF
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1047455) | | Descriptor: | 14-3-3 protein sigma, PHE-PRO-ALA-TPO-VAL, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]-2-(4-chloranylphenoxy)-2-methyl-propanamide | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BXO
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1075287) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]-4,4-bis(fluoranyl)-1-phenoxy-cyclohexane-1-carboxamide | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-12-09 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BXS
 
 | |
8BXQ
 
 | |
8BY9
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1075292) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]-1-(4-chloranylphenoxy)-4,4-bis(fluoranyl)cyclohexane-1-carboxamide | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
