5VEA
 
 | |
5VED
 
 | |
6SSO
 
 | | EDN mutant L45H | | Descriptor: | ACETATE ION, Non-secretory ribonuclease | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and functional characterization of new family enzymes derivates from human RNase 1 and 3 with antimicrobial and ribonuclease activity
To Be Published
|
|
6E5R
 
 | |
5A0F
 
 | | Crystal structure of Yersinia Afp18-modified RhoA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Jank, T, Schimpl, M, van Aalten, D.M. | | Deposit date: | 2015-04-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tyrosine glycosylation of Rho by Yersinia toxin impairs blastomere cell behaviour in zebrafish embryos.
Nat Commun, 6, 2015
|
|
4ZSP
 
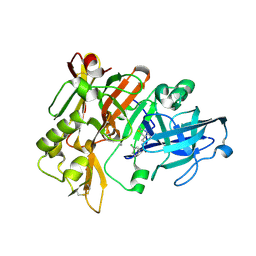 | | BACE crystal structure with bicyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(4aS,6S,8aR)-2-amino-4a,5,6,7,8,8a-hexahydro-4H-3,1-benzothiazin-6-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4PEP
 
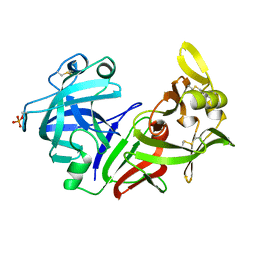 | |
4ZTS
 
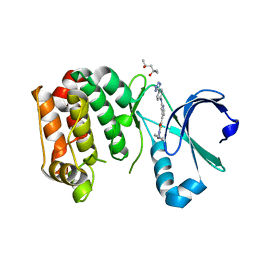 | | Human Aurora A catalytic domain bound to FK1142 | | Descriptor: | (2Z,5Z)-2-[(4-ethylphenyl)imino]-3-methyl-5-[(2-{[4-(1H-tetrazol-5-yl)phenyl]amino}pyridin-4-yl)methylidene]-1,3-thiazolidin-4-one, (4S)-2-METHYL-2,4-PENTANEDIOL, Aurora kinase A | | Authors: | Marcaida, M.J, Kilchmann, F, Schick, T, Reymond, J.L. | | Deposit date: | 2015-05-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Selective Aurora A Kinase Inhibitor by Virtual Screening.
J.Med.Chem., 59, 2016
|
|
6SDW
 
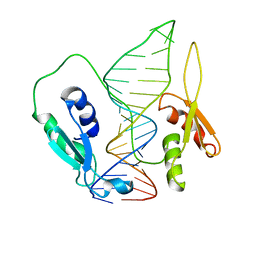 | |
4PRF
 
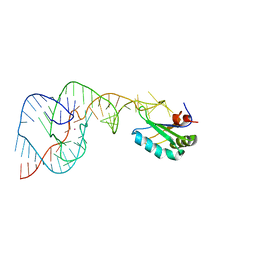 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
4PRU
 
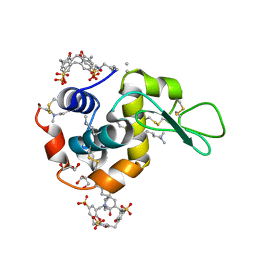 | | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 2.2 A resolution | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, GLYCEROL, Lysozyme C | | Authors: | McGovern, R.E, Lyons, J.A, Crowley, P.B. | | Deposit date: | 2014-03-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Chem Sci, 6, 2015
|
|
8WF5
 
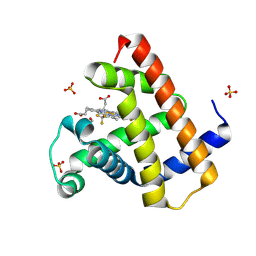 | | Horse heart myoglobin reconstituted with an iron complex of porphyrin bearing two CF3 groups (rMb(FePor(CF3)2)) | | Descriptor: | GLYCEROL, Myoglobin, PROTOPORPHYRIN(CF3)2 CONTAINING FE, ... | | Authors: | Kagawa, Y, Oohora, K, Himiyama, T, Suzuki, A, Hayashi, T. | | Deposit date: | 2023-09-19 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Redox Engineering of Myoglobin by Cofactor Substitution to Enhance Cyclopropanation Reactivity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6SNJ
 
 | |
5VFM
 
 | |
6E7J
 
 | | HIV-1 wild type protease with GRL-042-17A, 3-phenylhexahydro-2h-cyclopenta[d]oxazol-2-one with a bicyclic oxazolidinone scaffold as the P2 ligand | | Descriptor: | (3aS,5R,6aR)-2-oxo-3-phenylhexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
6T28
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6E8C
 
 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | Authors: | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
6T29
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6E8I
 
 | |
4PHI
 
 | | Crystal structure of HEWL with hexatungstotellurate(VI) | | Descriptor: | 6-tungstotellurate(VI), ACETATE ION, GLYCEROL, ... | | Authors: | Bijelic, A, Molitor, C, Mauracher, S.G, Al-Oweini, R, Kortz, U, Rompel, A. | | Deposit date: | 2014-05-06 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Hen Egg-White Lysozyme Crystallisation: Protein Stacking and Structure Stability Enhanced by a Tellurium(VI)-Centred Polyoxotungstate.
Chembiochem, 16, 2015
|
|
6E99
 
 | |
5VIL
 
 | | Crystal structure of ASK1 kinase domain with a potent inhibitor (analog 6) | | Descriptor: | 2-methoxy-N-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-5-sulfamoylbenzamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Jasti, J, Chang, J, Kurumbail, R. | | Deposit date: | 2017-04-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Rational approach to highly potent and selective apoptosis signal-regulating kinase 1 (ASK1) inhibitors.
Eur J Med Chem, 145, 2017
|
|
4WYZ
 
 | | The crystal structure of the A109G mutant of RNase A in complex with 3'UMP | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | French, R.L, Gagne, D, Doucet, N, Simonovic, M. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Perturbation of the Conformational Dynamics of an Active-Site Loop Alters Enzyme Activity.
Structure, 23, 2015
|
|
4PJM
 
 | | Myosin VI motor domain in the Pi release state, space group P212121 - soaked with PO4 - located in the active site | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
6EGY
 
 | | Crystal structure of cytochrome c in complex with mono-PEGylated sulfonatocalix[4]arene | | Descriptor: | Cytochrome c iso-1, HEME C, SULFATE ION, ... | | Authors: | Mummidivarapu, V.V.S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent PEGylation via Sulfonatocalix[4]arene-A Crystallographic Proof.
Bioconjug.Chem., 29, 2018
|
|
