1FVI
 
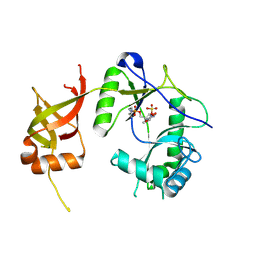 | | CRYSTAL STRUCTURE OF CHLORELLA VIRUS DNA LIGASE-ADENYLATE | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORELLA VIRUS DNA LIGASE-ADENYLATE, SULFATE ION | | Authors: | Odell, M, Sriskanda, V, Shuman, S, Nikolov, D.B. | | Deposit date: | 2000-09-20 | | Release date: | 2000-11-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining.
Mol.Cell, 6, 2000
|
|
2Q2U
 
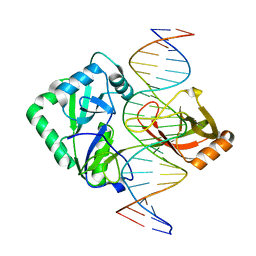 | | Structure of Chlorella virus DNA ligase-product DNA complex | | Descriptor: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*CP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', Chlorella virus DNA ligase | | Authors: | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2Q2T
 
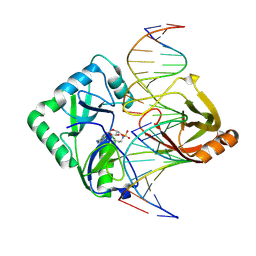 | | Structure of Chlorella virus DNA ligase-adenylate bound to a 5' phosphorylated nick | | Descriptor: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', 5'-D(P*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', ... | | Authors: | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1A0I
 
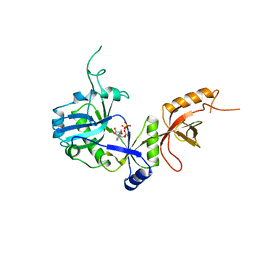 | | ATP-DEPENDENT DNA LIGASE FROM BACTERIOPHAGE T7 COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA LIGASE | | Authors: | Subramanya, H.S, Doherty, A.J, Ashford, S.R, Wigley, D.B. | | Deposit date: | 1997-12-01 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7.
Cell(Cambridge,Mass.), 85, 1996
|
|
3L2P
 
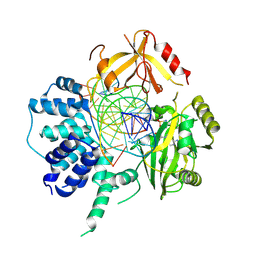 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
6KJM
 
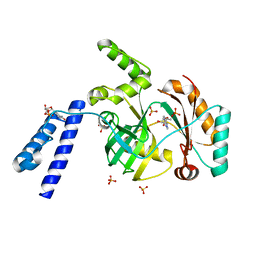 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based identification of first-in-class fragment inhibitors that target the NMN pocket of M. tuberculosis NAD + -dependent DNA ligase A.
J.Struct.Biol., 213, 2021
|
|
3UQ8
 
 | |
8U6X
 
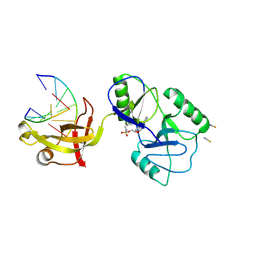 | | ATP-dependent DNA ligase Lig E from Neisseria gonorrhoeae | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*CP*CP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Williamson, A, Pan, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A role for the ATP-dependent DNA ligase lig E of Neisseria gonorrhoeae in biofilm formation.
Bmc Microbiol., 24, 2024
|
|
6KDU
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3II6
 
 | |
4CC5
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
4CC6
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-{[2-(1H-pyrazolo[3,4-c]pyridin-3-yl)-6-(trifluoromethyl)pyridin-4-yl]amino}ethanol, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
6DT1
 
 | | Crystal structure of the ligase from bacteriophage T4 complexed with DNA intermediate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction.
Nucleic Acids Res., 46, 2018
|
|
1B04
 
 | |
3QVG
 
 | | XRCC1 bound to DNA ligase | | Descriptor: | DNA ligase 3, DNA repair protein XRCC1 | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
1VS0
 
 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
3PC7
 
 | |
8VDS
 
 | | DNA Ligase 1 with nick DNA 3'rG:C | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*G)-D(P*GP*TP*CP*GP*GP*AP*C)-3') | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VDT
 
 | | DNA Ligase 1 with nick DNA 3'rA:T | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*TP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*A)-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
6IMJ
 
 | | The crystal structure of Se-AsfvLIG:DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
3PC8
 
 | | X-ray crystal structure of the heterodimeric complex of XRCC1 and DNA ligase III-alpha BRCT domains. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA ligase 3, DNA repair protein XRCC1, ... | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2010-10-21 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
2LJ6
 
 | |
9BS4
 
 | | DNA Ligase 1 E346A/E592A double mutant with 5'-rG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | KanalElamparithi, B, Caglayan, M. | | Deposit date: | 2024-05-12 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of LIG1 uncover the mechanism of sugar discrimination against 5'-RNA-DNA junctions during ribonucleotide excision repair.
J.Biol.Chem., 300, 2024
|
|
7KR3
 
 | |
7KR4
 
 | |
