7TYX
 
 | | Human Amylin2 Receptor in complex with Gs and rat amylin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
6GP8
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112A in complex AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GQS
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with AMPCP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
8T3N
 
 | | Solution NMR structure of synthetic peptide AMPCry10Aa_5 rational designed from Cry10Aa bacterial protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | SOLUTION NMR | | Cite: | Anti-Staphy Peptides Rationally Designed from Cry10Aa Bacterial Protein.
Acs Omega, 9, 2024
|
|
1BWY
 
 | | NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN | | Descriptor: | PROTEIN (HEART FATTY ACID BINDING PROTEIN) | | Authors: | Lassen, D, Luecke, C, Kveder, M, Mesgarzadeh, A, Schmidt, J.M, Specht, B, Lezius, A, Spener, F, Rueterjans, H. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy.
Eur.J.Biochem., 230, 1995
|
|
1CNE
 
 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NITRATE REDUCTASE | | Authors: | Lu, G, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-02-01 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
1CNF
 
 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NITRATE REDUCTASE | | Authors: | Lu, G, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-02-01 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
1BMP
 
 | | BONE MORPHOGENETIC PROTEIN-7 | | Descriptor: | BONE MORPHOGENETIC PROTEIN-7 | | Authors: | Griffith, D.L, Scott, D.L. | | Deposit date: | 1995-12-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
6OR6
 
 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (tail region) | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
8OTW
 
 | | Cryo-EM structure of Strongylocentrotus purpuratus SLC9C1 in presence of cAMP | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
6OR5
 
 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (ring region) | | Descriptor: | Midasin, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
8TW8
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA complex in Apo state conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
8TW7
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA complex in Apo state conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
8TWB
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
8P9Y
 
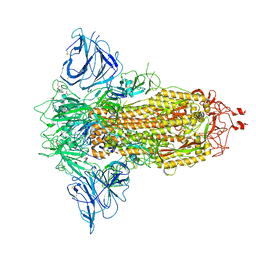 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8P99
 
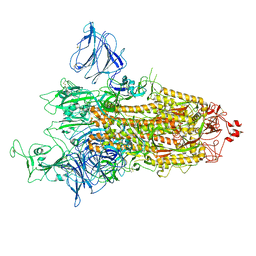 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
7Z38
 
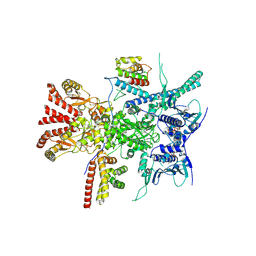 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z37
 
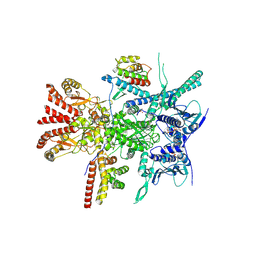 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
6S8M
 
 | | S. pombe microtubule decorated with Cut7 motor domain in the AMPPNP state | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Moores, C.A, von Loeffelholz, O. | | Deposit date: | 2019-07-10 | | Release date: | 2019-08-21 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM Structure (4.5- angstrom ) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance.
J.Mol.Biol., 431, 2019
|
|
8TW9
 
 | | Cryo-EM structure of S. cerevisiae PolE-Ctf18-8-1-DNA | | Descriptor: | Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
8TWA
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-PolE-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM reveals that Ctf18-RFC collaborates with PolE to load the PCNA clamp onto a leading strand DNA
To Be Published
|
|
8C89
 
 | | SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab) | | Descriptor: | 17T2 Fab heavy chain, 17T2 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Modrego, A, Carlero, D, Bueno-Carrasco, M.T, Santiago, C, Carolis, C, Arranz, R, Blanco, J, Magri, G. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | A monoclonal antibody targeting a large surface of the receptor binding motif shows pan-neutralizing SARS-CoV-2 activity.
Nat Commun, 15, 2024
|
|
9BKQ
 
 | | Structure of penguinpox cGAMP PDE in apo and post reaction states | | Descriptor: | 9-{3-O-[(S)-thiophosphono]-alpha-L-lyxofuranosyl}-9H-purin-6-amine, GUANINE, Penguinpox cGAMP PDE | | Authors: | Hobbs, S.J, Nomburg, J, Doudna, J.A, Kranzusch, P.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Animal and bacterial viruses share conserved mechanisms of immune evasion
Cell(Cambridge,Mass.), 2024
|
|
