5Q0C
 
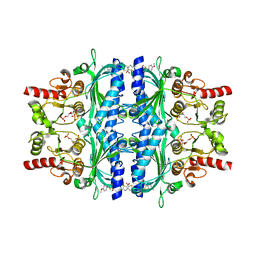 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with fructose-2,6-diphosphate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(4-methoxyphenyl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with f2,6p
To be published
|
|
2CMR
 
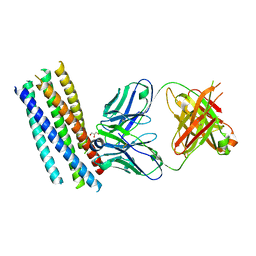 | | Crystal structure of the HIV-1 neutralizing antibody D5 Fab bound to the gp41 inner-core mimetic 5-helix | | Descriptor: | D5, GLYCEROL, TRANSMEMBRANE GLYCOPROTEIN | | Authors: | Luftig, M.A, Mattu, M, Di Giovine, P, Geleziunas, R, Hrin, R, Barbato, G, Bianchi, E, Miller, M.D, Pessi, A, Carfi, A. | | Deposit date: | 2006-05-11 | | Release date: | 2006-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for HIV-1 Neutralization by a Gp41 Fusion Intermediate-Directed Antibody
Nat.Struct.Mol.Biol., 13, 2006
|
|
2IEG
 
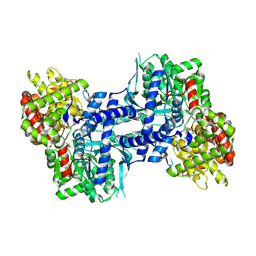 | | Crystal structure of rabbit muscle glycogen phosphorylase in complex with 3,4-dihydro-2-quinolone | | Descriptor: | (2S)-N-[(3S)-1-(2-AMINO-2-OXOETHYL)-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-3-YL]-2-CHLORO-2H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Glycogen phosphorylase, ... | | Authors: | Birch, A.M, Kenny, P.W, Oikonomakos, N.G, Otterbein, L, Schofield, P, Whittamore, P.R.O, Whalley, D.P, Rowsell, S, Pauptit, R, Pannifer, A, Breed, J, Minshull, C. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of potent, orally active 1-substituted-3,4-dihydro-2-quinolone glycogen phosphorylase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2AGD
 
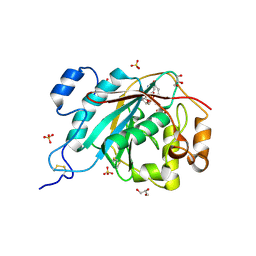 | | Crystal Structure of Human M340H-Beta-1,4-Galactosyltransferase-I(M340H-B4Gal-T1) in Complex with GlcNAc-beta1,4-Man-alpha1,3-Man-beta-OR | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
2FHY
 
 | |
5Q04
 
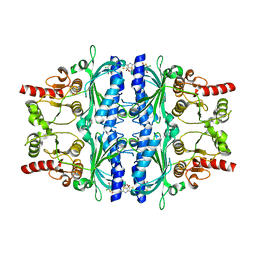 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea | | Descriptor: | 4-bromo-N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-chlorothiophene-2-sulfonamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea
To be published
|
|
5Q05
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methoxyethyl)-4-methylthiophen-2-yl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(2-methoxyethyl)-4-methylthiophen-2-yl]sulfonylurea
To be published
|
|
2FBP
 
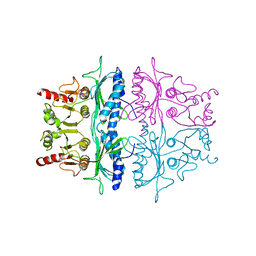 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | Deposit date: | 1990-06-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
2I51
 
 | |
4GZJ
 
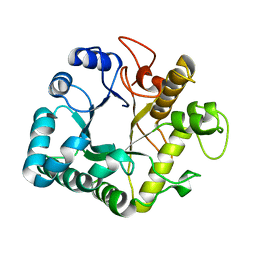 | | Active-site mutant of potato endo-1,3-beta-glucanase in complex with laminaratriose and laminaratetrose | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2012-09-06 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of an active-site mutant of a plant 1,3-beta-glucanase in complex with oligosaccharide products of hydrolysis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2FIE
 
 | |
2FIX
 
 | |
2FK5
 
 | | Crystal structure of l-fuculose-1-phosphate aldolase from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, SULFATE ION, fuculose-1-phosphate aldolase | | Authors: | Jeyakanthan, J, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-04 | | Release date: | 2007-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary X-ray crystallographic study of the L-fuculose-1-phosphate aldolase (FucA) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2HQ9
 
 | |
3OHH
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (bace-wt) complex with bms-681889 aka n~1~-butyl-5-cyano- n~3~-((1s,2r)-1-(3,5-difluorobenzyl)-2-hydroxy-3-((3- methoxybenzyl)amino)propyl)-n~1~-methyl-1h-indole-1,3- dicarboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, N~1~-butyl-5-cyano-N~3~-{(1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-N~1~-methyl-1H-indole-1,3-dicarboxamide, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2010-08-17 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthesis and SAR of indole-and 7-azaindole-1,3-dicarboxamide hydroxyethylamine inhibitors of BACE-1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2FD0
 
 | | HIV-1 DIS kissing-loop in complex with lividomycin | | Descriptor: | (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, CHLORIDE ION, HIV-1 DIS RNA, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
2FUA
 
 | | L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T WITH COBALT | | Descriptor: | BETA-MERCAPTOETHANOL, COBALT (II) ION, L-FUCULOSE-1-PHOSPHATE ALDOLASE, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined high-resolution structure of the metal-ion dependent L-fuculose-1-phosphate aldolase (class II) from Escherichia coli.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3RUB
 
 | | CRYSTAL STRUCTURE OF THE UNACTIVATED FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM TOBACCO REFINED AT 2.0-ANGSTROMS RESOLUTION | | Descriptor: | ASPARAGINE, RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, FORM III, ... | | Authors: | Schreuder, H, Cascio, D, Curmi, P.M.G, Chapman, M.S, Suh, S.W, Eisenberg, D.S. | | Deposit date: | 1990-05-25 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution.
J.Biol.Chem., 267, 1992
|
|
2HOT
 
 | | Phage selected homeodomain bound to modified DNA | | Descriptor: | 3-PROP-2-YN-1-YL-1,3-OXAZOLIDIN-2-ONE, 5'-D(*AP*TP*CP*CP*GP*GP*GP*GP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*CP*CP*CP*CP*GP*GP*A)-3', ... | | Authors: | Feldman, M.E, Simon, M.D, Shokat, K.M. | | Deposit date: | 2006-07-16 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure and properties of a re-engineered homeodomain protein-DNA interface.
Acs Chem.Biol., 1, 2006
|
|
4ACX
 
 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 23 | | Descriptor: | (8R)-8-[4-(DIFLUOROMETHOXY)PHENYL]-3,3-DIFLUORO-8-[3-(3-METHOXYPROP-1-YN-1-YL)PHENYL]-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
3N9K
 
 | | F229A/E292S Double Mutant of Exo-beta-1,3-glucanase from Candida albicans in Complex with Laminaritriose at 1.7 A | | Descriptor: | CALCIUM ION, Glucan 1,3-beta-glucosidase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2010-05-30 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance
Febs J., 277, 2010
|
|
1PEZ
 
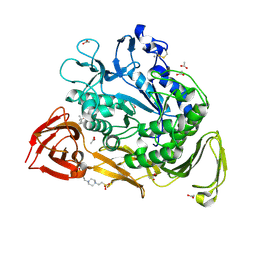 | | Bacillus circulans strain 251 mutant A230V | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETIC ACID, ... | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Conversion of Cyclodextrin Glycosyltransferase into a Starch Hydrolase by Directed Evolution: The Role of Alanine 230 in Acceptor Subsite +1
Biochemistry, 42, 2003
|
|
4JM2
 
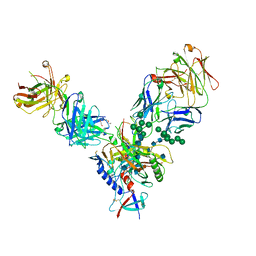 | |
3OHF
 
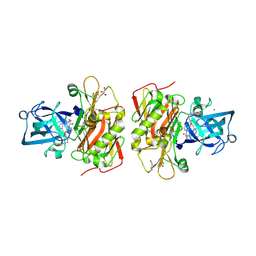 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with bms-655295 aka n~3~-((1s,2r)-1- benzyl-2-hydroxy-3-((3-methoxybenzyl)amino)propyl)-n~1~, n~1~-dibutyl-1h-indole-1,3-dicarboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2010-08-17 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and SAR of indole-and 7-azaindole-1,3-dicarboxamide hydroxyethylamine inhibitors of BACE-1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2G01
 
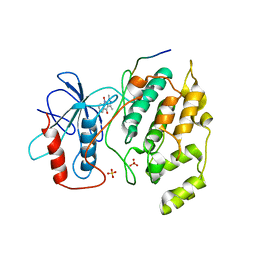 | | Pyrazoloquinolones as Novel, Selective JNK1 inhibitors | | Descriptor: | 6-CHLORO-9-HYDROXY-1,3-DIMETHYL-1,9-DIHYDRO-4H-PYRAZOLO[3,4-B]QUINOLIN-4-ONE, C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-02-10 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Synthesis and SAR of 1,9-dihydro-9-hydroxypyrazolo[3,4-b]quinolin-4-ones as novel, selective c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
