2GMK
 
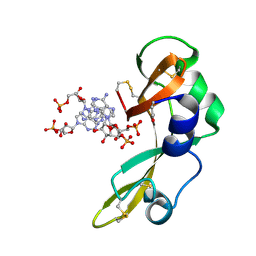 | | Crystal structure of onconase double mutant with spontaneously-assembled (AMP) 4 stack | | Descriptor: | ADENOSINE MONOPHOSPHATE, P-30 protein | | Authors: | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
4U8Z
 
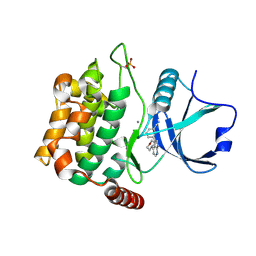 | | Crystal structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06447475) | | Descriptor: | 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, MANGANESE (II) ION, Serine/threonine-protein kinase 24 | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
2GPE
 
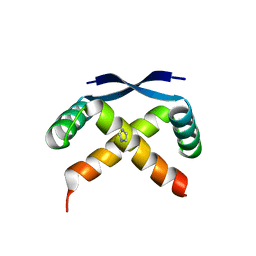 | |
4TVH
 
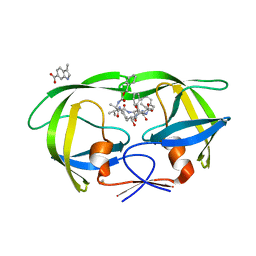 | | HIV Protease (PR) dimer in closed form with TL-3 in active site and fragment AK-2097 in the outside/top of flap | | Descriptor: | 3-(morpholin-4-ylmethyl)-1H-indole-6-carboxylic acid, BETA-MERCAPTOETHANOL, Protease, ... | | Authors: | Tiefenbrunn, T, Kislukhin, A, Finn, M.G, Stout, C.D. | | Deposit date: | 2014-06-26 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease.
J.Phys.Chem.B, 119, 2015
|
|
3BMC
 
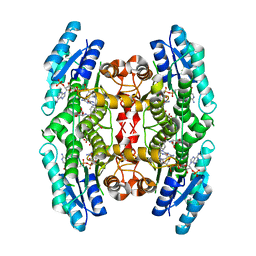 | |
8FCC
 
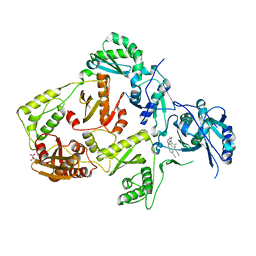 | | HIV-1 Reverse Transcriptase in complex with 5-membered bicyclic core NNRTI | | Descriptor: | 4-[(9-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-8-oxo-8,9-dihydro-7H-purin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
4TXJ
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with thymidine | | Descriptor: | SULFATE ION, THYMIDINE, Uridine phosphorylase | | Authors: | Torini, J, Marinho, A, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
8FCD
 
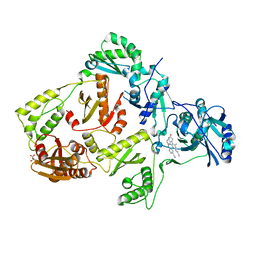 | | HIV-1 Reverse Transcriptase in complex with 6-membered bicyclic core NNRTI | | Descriptor: | 4-[(8-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-6-oxo-5,6,7,8-tetrahydropteridin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
8FCE
 
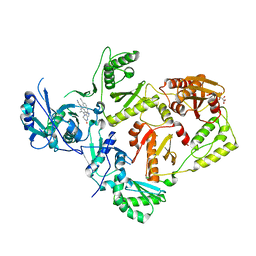 | | HIV-1 Reverse Transcriptase in complex with 7-membered bicyclic core NNRTI | | Descriptor: | 4-[(9-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
8G08
 
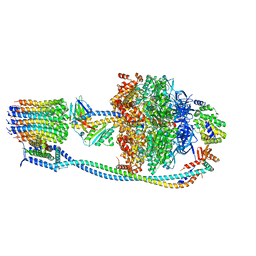 | |
8PKO
 
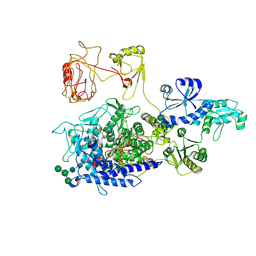 | | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Hitchman, C.J, Lia, A, Bayo, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer).
To Be Published
|
|
2H89
 
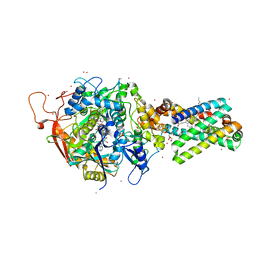 | | Avian Respiratory Complex II with Malonate Bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
2ABS
 
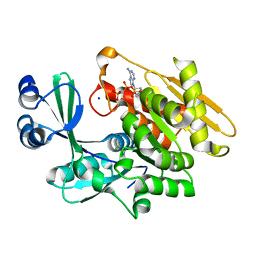 | | Crystal structure of T. gondii adenosine kinase complexed with AMP-PCP | | Descriptor: | CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-16 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of Toxoplasma gondii adenosine kinase in complex with an ATP analog at 1.1 angstroms resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4U5J
 
 | | C-Src in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Chen, Y, Duan, Y, Chen, L. | | Deposit date: | 2014-07-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | c-Src Binds to the Cancer Drug Ruxolitinib with an Active Conformation
Plos One, 9, 2014
|
|
8G09
 
 | |
2ADU
 
 | | Human Methionine Aminopeptidase Complex with 4-Aryl-1,2,3-triazole Inhibitor | | Descriptor: | 4-(3-METHYLPHENYL)-1H-1,2,3-TRIAZOLE, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Kallander, L.S, Lu, Q, Chen, W, Tomaszek, T, Yang, G, Tew, D, Meek, T.D, Hofmann, G.A, Schulz-Pritchard, C.K, Smith, W.W, Janson, C.A, Ryan, M.D, Zhang, G.F, Johanson, K.O, Kirkpatrick, R.B, Ho, T.F, Fisher, P.W, Mattern, M.R, Johnson, R.K, Hansbury, M.J, Winkler, J.D, Ward, K.W, Veber, D.F, Thompson, S.K. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized To Inhibit Angiogenesis in Vivo
J.Med.Chem., 48, 2005
|
|
8G0B
 
 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase FO region | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ATP synthase subunit a, ATP synthase subunit b, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
3PVW
 
 | |
8FLN
 
 | | Crystal structure of BTK C481S kinase domain in complex with pirtobrutinib | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
4U61
 
 | |
4U9D
 
 | |
4M2U
 
 | | Carbonic Anhydrase II in complex with Dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Pinard, M.P, Boone, C.D, Rife, B.D, Supuran, C.T, Mckenna, R. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases.
Bioorg.Med.Chem., 21, 2013
|
|
8G0C
 
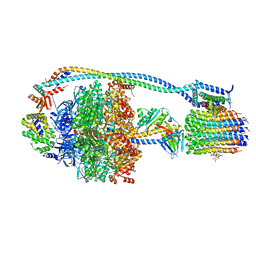 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8FLL
 
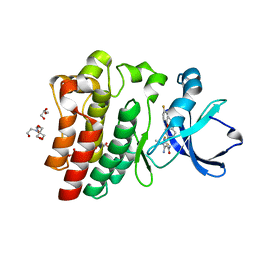 | | Crystal structure of BTK kinase domain in complex with pirtobrutinib | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
8G0E
 
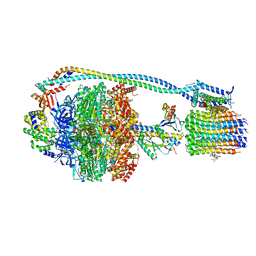 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
