3V4I
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and AZTTP | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7983 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3QGF
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4OOZ
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose | | Descriptor: | Beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, ... | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
1R0A
 
 | | Crystal structure of HIV-1 reverse transcriptase covalently tethered to DNA template-primer solved to 2.8 angstroms | | Descriptor: | 5'-D(*A*TP*GP*CP*AP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*GP*GP*T)-3', 5'-D(*C*CP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*AP*GP*CP*GP*CP*CP*GP*(2DA))-3', GLYCEROL, ... | | Authors: | Tuske, S, Ding, J, Arnold, E. | | Deposit date: | 2003-09-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nonnucleoside inhibitor binding affects the interactions of the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase with DNA.
J.Virol., 78, 2004
|
|
3QGG
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2DQZ
 
 | | Crystal structure of human carboxylesterase in complex with homatropine, coenzyme A, and palmitate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COENZYME A, FLUORIDE ION, ... | | Authors: | Bencharit, S, Redinbo, M.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1
J.Mol.Biol., 363, 2006
|
|
3FBP
 
 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | Deposit date: | 1990-06-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
1BIS
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BWB
 
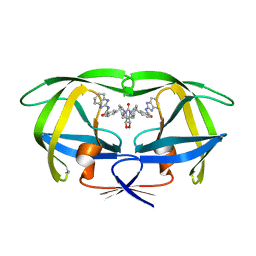 | | HIV-1 PROTEASE (V82F/I84V) DOUBLE MUTANT COMPLEXED WITH SD146 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6ALPHA,7ALPHA)]-3,3'-{{TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-DIYL]BIS(METHYLENE)]BIS[N-1H-BENZIMIDAZOL-2-YLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1IZ8
 
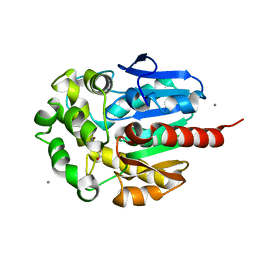 | | Re-refinement of the structure of hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Streltsov, V.A. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
3F5E
 
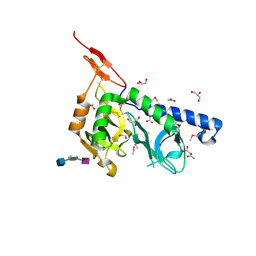 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 2'F-3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
4BHI
 
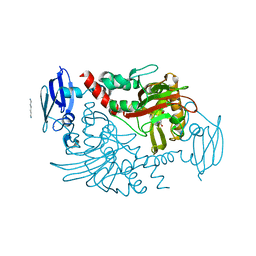 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-(1,1,1,2-Tetramethylhydrazin-1-ium-2-yl)propanoate | | Descriptor: | 3-(1,1,1,2-TETRAMETHYLHYDRAZIN-1-IUM-2-YL)PROPANOATE, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-04-03 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
3F5A
 
 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
3DS0
 
 | |
3D5Z
 
 | | Crystal Structure Analysis of 1,5-alpha-arabinanase catalytic mutant (AbnBE201A) complexed to arabinotriose | | Descriptor: | CALCIUM ION, Intracellular arabinanase, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-beta-L-arabinofuranose | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-05-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
1GWW
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - ALPHA-D-GLUCOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GZ7
 
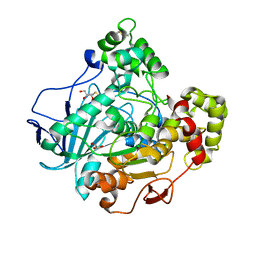 | | Crystal structure of the closed state of lipase 2 from Candida rugosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LIPASE 2 | | Authors: | Mancheno, J.M, Hermoso, J.A. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights Into the Lipase/Esterase Behavior in the Candida Rugosa Lipases Family: Crystal Structure of the Lipase 2 Isoenzyme at 1.97A Resolution
J.Mol.Biol., 332, 2003
|
|
3PZQ
 
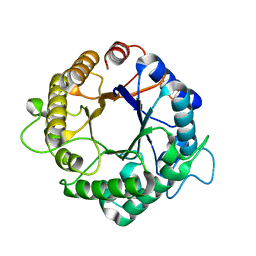 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol | | Descriptor: | Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3DNO
 
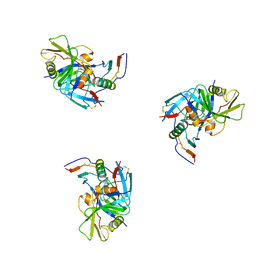 | | Molecular structure for the HIV-1 gp120 trimer in the CD4-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
1RUS
 
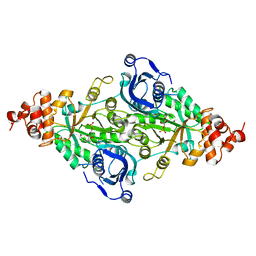 | | CRYSTAL STRUCTURE OF THE BINARY COMPLEX OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE AND ITS PRODUCT, 3-PHOSPHO-D-GLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1991-10-10 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the binary complex of ribulose-1,5-bisphosphate carboxylase and its product, 3-phospho-D-glycerate.
J.Biol.Chem., 264, 1989
|
|
3DS2
 
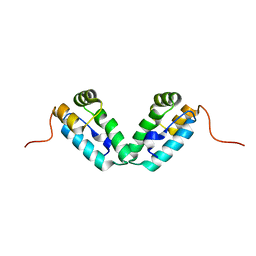 | | HIV-1 capsid C-terminal domain mutant (Y169A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Vaney, M.-C, Igonet, S, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
3DNN
 
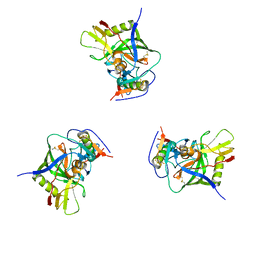 | | Molecular structure for the HIV-1 gp120 trimer in the unliganded state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
3DS5
 
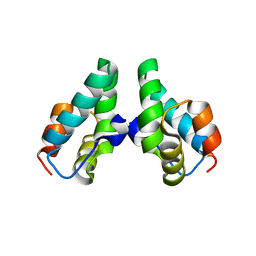 | | HIV-1 capsid C-terminal domain mutant (N183A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
2VJ6
 
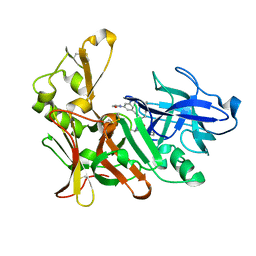 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-(ethylamino)-5-(2-oxo-1-pyrrolidinyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-{[(1S)-2-(cyclohexylamino)-1-methyl-2-oxoethyl]amino}-2-hydroxypropyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3WH1
 
 | | Crystal Structure of a Family GH19 Chitinase from Bryum coronatum in complex with (GlcNAc)4 at 1.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2013-08-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of a "loopless" GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center.
Biochim.Biophys.Acta, 1844, 2014
|
|
