2H3W
 
 | | Crystal structure of the S554A/M564G mutant of murine carnitine acetyltransferase in complex with hexanoylcarnitine and CoA | | Descriptor: | (R)-3-CARBOXY-2-(HEXANOYLOXY)-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, COENZYME A, carnitine acetyltransferase | | Authors: | Hsiao, Y.S, Jogl, G, Tong, L. | | Deposit date: | 2006-05-23 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of murine carnitine acetyltransferase in ternary complexes with its substrates
J.Biol.Chem., 281, 2006
|
|
4CDN
 
 | | Crystal structure of M. mazei photolyase with its in vivo reconstituted 8-HDF antenna chromophore | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
2V4E
 
 | | A non-cytotoxic DsRed variant for whole-cell labeling | | Descriptor: | RED FLUORESCENT PROTEIN DRFP583 | | Authors: | Strack, R.L, Strongin, D.E, Bhattacharyya, D, Tao, W, Berman, A, Broxmeyer, H.E, Keenan, R.J, Glick, B.S. | | Deposit date: | 2008-09-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Noncytotoxic Dsred Variant for Whole-Cell Labeling.
Nat.Methods, 5, 2008
|
|
4CDM
 
 | | Crystal structure of M. mazei photolyase soaked with synthetic 8-HDF | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
4BVS
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, CYANURIC ACID AMIDOHYDROLASE, MAGNESIUM ION | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
3VT8
 
 | | Crystal structures of rat VDR-LBD with W282R mutation | | Descriptor: | (1R,3R,7E,9beta,17beta)-9-butyl-17-[(2R)-6-hydroxy-6-methylheptan-2-yl]-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
4B36
 
 | |
1QKQ
 
 | |
3UYC
 
 | | Designed protein KE59 R8_2/7A | | Descriptor: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VT4
 
 | | Crystal structures of rat VDR-LBD with R270L mutation | | Descriptor: | (1R,2Z,3R,5E,7E)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethylidene)-9,10-secoestra-5,7,16-triene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
4CJC
 
 | | orthorhombic crystal form of Bogt6a E192Q in complex with UDP-GalNAc, UDP, GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSYLTRANSFERASE FAMILY 6, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Pham, T, Stinson, B, Thiyagarajan, N, Lizotte-Waniewski, M, Brew, K, Acharya, K.R. | | Deposit date: | 2013-12-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structures of Complexes of a Metal-Independent Glycosyltransferase Gt6 from Bacteroides Ovatus with Udp-Galnac and its Hydrolysis Products.
J.Biol.Chem., 289, 2014
|
|
2V8T
 
 | | Crystal structure of Mn catalase from Thermus Thermophilus complexed with chloride | | Descriptor: | CHLORIDE ION, LITHIUM ION, MANGANESE (II) ION, ... | | Authors: | Antonyuk, S.V, Barynin, V.V, Vaguine, A.A, Melik-Adamyan, W.R, Popov, A.N, Lamsin, V.S, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Three-Dimentional Structure of the Enzyme Dimanganese Catalase from Thermus Thermophilus at 1 Angstrom Resolution
Crystallogr.Rep.(Transl. Kristallografiya), 45, 2000
|
|
2SKE
 
 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH PHOSPHITE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | INOSINIC ACID, PHOSPHITE ION, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
2SKC
 
 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH FLUOROPHOSPHATE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | FLUORO-PHOSPHITE ION, INOSINIC ACID, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
4CJB
 
 | | orthorhombic crystal form of Bogt6a E192Q in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSYLTRANSFERASE FAMILY 6 | | Authors: | Pham, T, Stinson, B, Thiyagarajan, N, Lizotte-Waniewski, M, Brew, K, Acharya, K.R. | | Deposit date: | 2013-12-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structures of Complexes of a Metal-Independent Glycosyltransferase Gt6 from Bacteroides Ovatus with Udp-Galnac and its Hydrolysis Products.
J.Biol.Chem., 289, 2014
|
|
4CJ8
 
 | | monoclinic crystal form of Bogt6a E192Q in complex with UDP-GalNAc, UDP and GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, GLYCEROL, ... | | Authors: | Pham, T, Stinson, B, Thiyagarajan, N, Lizotte-Waniewski, M, Brew, K, Acharya, K.R. | | Deposit date: | 2013-12-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of Complexes of a Metal-Independent Glycosyltransferase Gt6 from Bacteroides Ovatus with Udp-Galnac and its Hydrolysis Products
J.Biol.Chem., 289, 2014
|
|
4D8O
 
 | |
2SKD
 
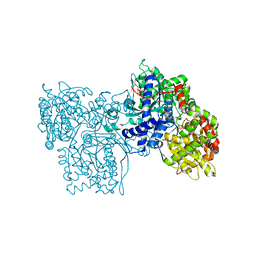 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH PHOSPHATE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | INOSINIC ACID, PHOSPHATE ION, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
2O86
 
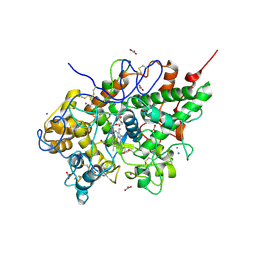 | | Crystal structure of a ternary complex of buffalo lactoperoxidase with nitrate and iodide at 2.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Sheikh, I.A, Singh, N, Singh, A.K, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-12-12 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a ternary complex of buffalo lactoperoxidase with nitrate and iodide at 2.8 A resolution
To be Published
|
|
2VJ0
 
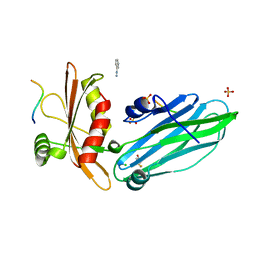 | | Crystal structure of the alpha-adaptin appendage domain, from the AP2 adaptor complex, in complex with an FXDNF peptide from amphiphysin1 and a WVXF peptide from synaptojanin P170 | | Descriptor: | AMPHIPHYSIN, AP-2 COMPLEX SUBUNIT ALPHA-2, BENZAMIDINE, ... | | Authors: | Ford, M.G.J, Praefcke, G.J.K, McMahon, H.T. | | Deposit date: | 2007-12-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Solitary and Repetitive Binding Motifs for the Ap2 Complex {Alpha}-Appendage in Amphiphysin and Other Accessory Proteins.
J.Biol.Chem., 283, 2008
|
|
2VYA
 
 | | Crystal Structure of fatty acid amide hydrolase conjugated with the drug-like inhibitor PF-750 | | Descriptor: | 4-(quinolin-3-ylmethyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Johnson, D.S, Wang, Z, Everdeen, D.S, Liimatta, M, Pabst, B, Bhattacharya, K, Nugent, R.A, Kamtekar, S, Cravatt, B.F, Ahn, K, Stevens, R.C. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Guided Inhibitor Design for Human Faah by Interspecies Active Site Conversion.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2W5O
 
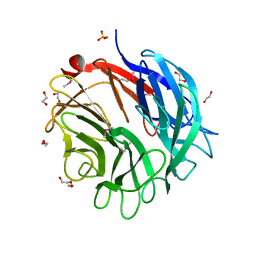 | | Complex structure of the GH93 alpha-L-arabinofuranosidase of Fusarium graminearum with arabinobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carapito, R, Imberty, A, Jeltsch, J.M, Byrns, S.C, Tam, P.H, Lowary, T.L, Varrot, A, Phalip, V. | | Deposit date: | 2008-12-11 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Basis of Arabinobio-Hydrolase Activity in Phytopathogenic Fungi. Crystal Structure and Catalytic Mechanism of Fusarium Graminearum Gh93 Exo-Alpha-L-Arabinanase.
J.Biol.Chem., 284, 2009
|
|
2Y70
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF GM23, MUTANT G89D, AN EXAMPLE OF CATALYTIC MIGRATION FROM TIM TO THIAMIN PHOSPHATE SYNTHASE. | | Descriptor: | ACETATE ION, SULFATE ION, TRIOSE-PHOSPHATE ISOMERASE | | Authors: | Saab-Rincon, G, Olvera, L, Olvera, M, Rudino-Pinera, E, Soberon, X, Morett, E. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolutionary Walk between (Beta/Alpha)(8) Barrels: Catalytic Migration from Triosephosphate Isomerase to Thiamin Phosphate Synthase.
J.Mol.Biol., 416, 2012
|
|
2Y6Z
 
 | | Crystallographic structure of GM23 an example of Catalytic migration from TIM to thiamin phosphate synthase. | | Descriptor: | PYROPHOSPHATE 2-, THIAMIN PHOSPHATE, TRIOSE-PHOSPHATE ISOMERASE | | Authors: | Saab-Rincon, G, Olvera, L, Olvera, M, Rudino-Pinera, E, Soberon, X, Morett, E. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Walk between (Beta/Alpha)(8) Barrels: Catalytic Migration from Triosephosphate Isomerase to Thiamin Phosphate Synthase.
J.Mol.Biol., 416, 2012
|
|
5OLR
 
 | | Rhamnogalacturonan lyase | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Rhamnogalacturonan lyase, ... | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
