6C63
 
 | | Crystal Structure of the Mango-II Fluorescent Aptamer Bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, POTASSIUM ION, RNA (32-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.900028 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
4EFC
 
 | | Crystal Structure of Adenylosuccinate Lyase from Trypanosoma Brucei, Tb427tmp.160.5560 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, ... | | Authors: | Wernimont, A.K, Loppnau, P, Osman, K.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Robinson, D.A, Wyatt, P.G, Gilbert, I.H, Fairlamb, A.H, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Trypanosoma Brucei, Tb427tmp.160.5560
To be Published
|
|
5VKI
 
 | | Crystal structure of P[19] rotavirus VP8* complexed with mucin core 2 | | Descriptor: | GLYCEROL, Outer capsid protein VP4, SULFATE ION, ... | | Authors: | Xu, S, Liu, Y, Woodruff, A, Zhong, W, Jiang, X, Kennedy, M.A. | | Deposit date: | 2017-04-21 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of glycan specificity of P[19] VP8*: Implications for rotavirus zoonosis and evolution.
PLoS Pathog., 13, 2017
|
|
6C69
 
 | | Structure of glycolipid aGSA[12,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
5RZO
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434925 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5-chloro-1,2,3-thiadiazol-4-yl)methyl]morpholine, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
1SML
 
 | | METALLO BETA LACTAMASE L1 FROM STENOTROPHOMONAS MALTOPHILIA | | Descriptor: | PROTEIN (PENICILLINASE), ZINC ION | | Authors: | Ullah, J.H, Walsh, T.R, Taylor, I.A, Emery, D.C, Verma, C.S, Gamblin, S.J, Spencer, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution.
J.Mol.Biol., 284, 1998
|
|
6C6E
 
 | | Structure of glycolipid aGSA[26,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
4M7F
 
 | | Crystal structure of tetrameric fibrinogen-like recognition domain of FIBCD1 with bound ManNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Shrive, A.K, Greenhough, T.J, Holmskov, U. | | Deposit date: | 2013-08-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Tetrameric Fibrinogen-like Recognition Domain of Fibrinogen C Domain Containing 1 (FIBCD1) Protein.
J.Biol.Chem., 289, 2014
|
|
5R4P
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with NM450-1 | | Descriptor: | (5S)-5-methyl-5-[(1-phenyl-1H-1,2,3-triazol-4-yl)methyl]pyrrolidine-2,4-dione, ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, ... | | Authors: | Kidd, S.L, Mateu, N, Talon, R, Krojer, T, Aimon, A, Bradley, A.R, Fairhead, M, Diaz-Saez, L, Sore, H.F, Madin, A, Huber, K.V.M, von Delft, F, Spring, D.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | Descriptor: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
4U24
 
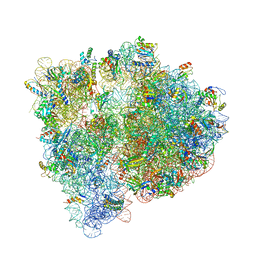 | | Crystal structure of the E. coli ribosome bound to dalfopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
6JAH
 
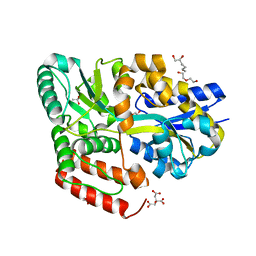 | | Crystal structure of ABC transporter alpha-glycoside-binding protein in complex with glucose | | Descriptor: | (2S)-heptane-1,2,7-triol, 1,2-ETHANEDIOL, ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
4CF0
 
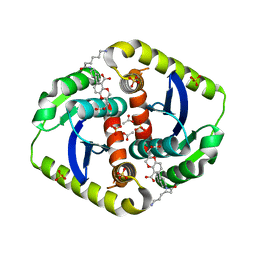 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2R)-6-[[(1R,2S)-2-(6-azanylhexanoylamino)-2,3-dihydro-1H-inden-1-yl]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
1SNP
 
 | |
4C1B
 
 | |
5RBQ
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library B06b | | Descriptor: | (1S)-1-(4-nitrophenyl)ethan-1-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1SNQ
 
 | |
4KC1
 
 | | Structure of the blood group glycosyltransferase AAglyB in complex with a pyridine inhibitor as a neutral pyrophosphate surrogate | | Descriptor: | 6-(1-beta-D-Glucopyranosyloxymethyl)-N-(5'-deoxyluridine-5'-yl)picolinamide, Fucosylglycoprotein alpha-N-acetylgalactosaminyltransferase, MANGANESE (II) ION, ... | | Authors: | Cuesta-Seijo, J.A, Wang, S, Lafont, D, Vidal, S, Palcic, M.M. | | Deposit date: | 2013-04-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of glycosyltransferase inhibitors: pyridine as a pyrophosphate surrogate.
Chemistry, 19, 2013
|
|
4C4X
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with C9 | | Descriptor: | 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6BQT
 
 | |
4EQC
 
 | | Crystal structure of PAK1 kinase domain in complex with FRAX597 inhibitor | | Descriptor: | 6-[2-chloro-4-(1,3-thiazol-5-yl)phenyl]-8-ethyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-04-18 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | FRAX597, a Small Molecule Inhibitor of the p21-activated Kinases, Inhibits Tumorigenesis of Neurofibromatosis Type 2 (NF2)-associated Schwannomas.
J.Biol.Chem., 288, 2013
|
|
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6JT3
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tadano, G, Komano, K, Yoshida, S, Suzuki, S, Nakahara, K, Fuchino, K, Fujimoto, K, Matsuoka, E, Yamamoto, T, Asada, N, Ito, H, Sakaguchi, G, Kanegawa, N, Kido, Y, Ando, S, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Bergh, A.V.D, Austin, N, Gijsen, H.J.M, Yamano, Y, Iso, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of an Extremely Potent Thiazine-Based beta-Secretase Inhibitor with Reduced Cardiovascular and Liver Toxicity at a Low Projected Human Dose.
J.Med.Chem., 62, 2019
|
|
1XC4
 
 | |
3BNY
 
 | | Crystal structure of aristolochene synthase complexed with 2-fluorofarnesyl diphosphate (2F-FPP) | | Descriptor: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Christianson, D.W. | | Deposit date: | 2007-12-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis
J.Biol.Chem., 283, 2008
|
|
