8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-2 RNA SL5 domain. | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
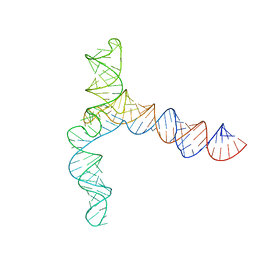 | | MERS 5' proximal stem-loop 5, conformation 3 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | Descriptor: | RNA (135-MER) | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2GU8
 
 | |
1DFC
 
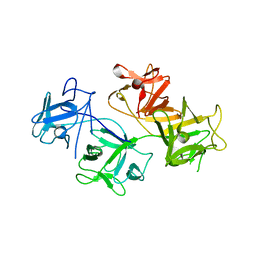 | | CRYSTAL STRUCTURE OF HUMAN FASCIN, AN ACTIN-CROSSLINKING PROTEIN | | Descriptor: | FASCIN | | Authors: | Fedorov, A.A, Fedorov, E.V, Ono, S, Matsumura, F, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-11-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein.
J.Mol.Biol., 400, 2010
|
|
1EES
 
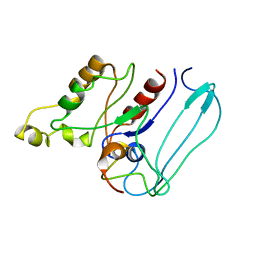 | | SOLUTION STRUCTURE OF CDC42HS COMPLEXED WITH A PEPTIDE DERIVED FROM P-21 ACTIVATED KINASE, NMR, 20 STRUCTURES | | Descriptor: | GTP-BINDING PROTEIN, P21-ACTIVATED KINASE | | Authors: | Gizachew, D, Guo, W, Chohan, K.C, Sutcliffe, M.J, Oswald, R.E. | | Deposit date: | 2000-02-02 | | Release date: | 2000-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of Cdc42Hs with a peptide derived from P-21 activated kinase.
Biochemistry, 39, 2000
|
|
2FAR
 
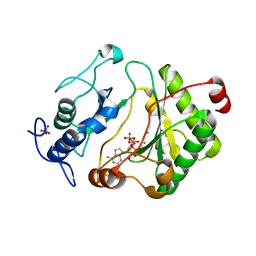 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with dATP and Manganese | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2KPK
 
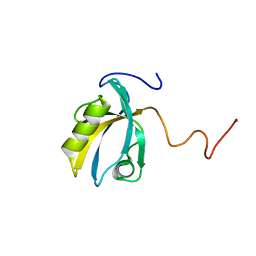 | | MAGI-1 PDZ1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
2KPL
 
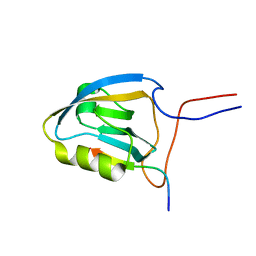 | | MAGI-1 PDZ1 / E6CT | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Protein E6 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
2HA4
 
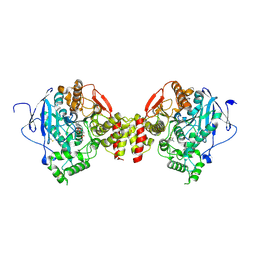 | | Crystal structure of mutant S203A of mouse acetylcholinesterase complexed with acetylcholine | | Descriptor: | ACETATE ION, ACETYLCHOLINE, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Radic, Z, Sulzenbacher, G, Kim, E, Taylor, P, Marchot, P. | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Substrate and product trafficking through the active center gorge of acetylcholinesterase analyzed by crystallography and equilibrium binding
J.Biol.Chem., 281, 2006
|
|
1DOA
 
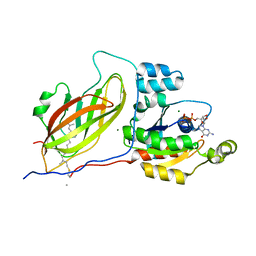 | | Structure of the rho family gtp-binding protein cdc42 in complex with the multifunctional regulator rhogdi | | Descriptor: | GERAN-8-YL GERAN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hoffman, G.R, Nassar, N, Cerione, R.C. | | Deposit date: | 1999-12-20 | | Release date: | 2000-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI.
Cell(Cambridge,Mass.), 100, 2000
|
|
2K1Z
 
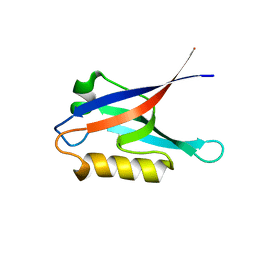 | |
2K3H
 
 | | Structural determinants for Ca2+ and PIP2 binding by the C2A domain of rabphilin-3A | | Descriptor: | CALCIUM ION, Rabphilin-3A | | Authors: | Coudevylle, N, Montaville, P, Leonov, A, Zweckstetter, M, Becker, S. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants for Ca2+ and Phosphatidylinositol 4,5-Bisphosphate Binding by the C2A Domain of Rabphilin-3A.
J.Biol.Chem., 283, 2008
|
|
2KQP
 
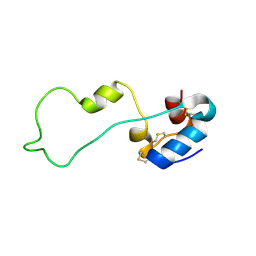 | |
5XVT
 
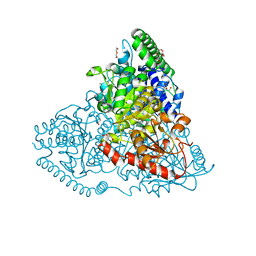 | | Crystal Structure of Transketolase in complex with hydroxylated TPP from Pichia Stipitis, crystal 2 | | Descriptor: | 2-[(2R)-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-oxidanyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Mesomeric Effect of Thiazolium on non-Kekule Diradicals in Pichia stipitis Transketolase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6O10
 
 | | E. coli cysteine desulfurase SufS | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Frantom, P.A. | | Deposit date: | 2019-02-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation of intermediates in the SufS cysteine desulfurase reaction reveals functional roles of conserved active-site residues.
J.Biol.Chem., 294, 2019
|
|
1U19
 
 | | Crystal Structure of Bovine Rhodopsin at 2.2 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Sugihara, M, Bondar, A.N, Elstner, M, Entel, P, Buss, V. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure
J.Mol.Biol., 342, 2004
|
|
3ZLV
 
 | | Crystal structure of mouse acetylcholinesterase in complex with tabun and HI-6 | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Artursson, E, Andersson, P.O, Akfur, C, Linusson, A, Borjegren, S, Ekstrom, F. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic-Site Conformational Equilibrium in Nerve-Agent Adducts of Acetylcholinesterase; Possible Implications for the Hi-6 Antidote Substrate Specificity.
Biochem.Pharmacol., 85, 2013
|
|
6O5V
 
 | | Binary complex of native hAChE with oxime reactivator RS-170B | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Productive reorientation of a bound oxime reactivator revealed in room temperature X-ray structures of native and VX-inhibited human acetylcholinesterase.
J.Biol.Chem., 294, 2019
|
|
6O69
 
 | |
