1PLW
 
 | |
1PRR
 
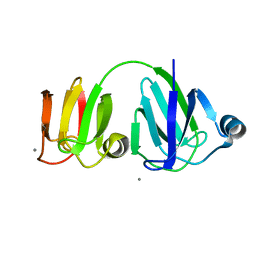 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
1PLX
 
 | |
1PRS
 
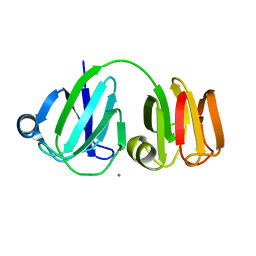 | | NMR-DERIVED THREE-DIMENSIONAL SOLUTION STRUCTURE OF PROTEIN S COMPLEXED WITH CALCIUM | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Bagby, S, Harvey, T.S, Eagle, S.G, Inouye, S, Ikura, M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR-derived three-dimensional solution structure of protein S complexed with calcium.
Structure, 2, 1994
|
|
1KR8
 
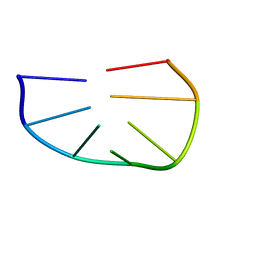 | |
1K1Z
 
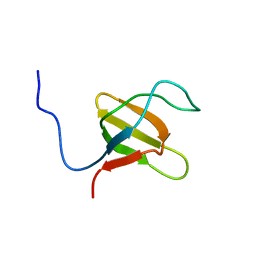 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
8QAJ
 
 | |
1RJJ
 
 | | Solution structure of a homodimeric hypothetical protein, At5g22580, a structural genomics target from Arabidopsis thaliana | | Descriptor: | expressed protein | | Authors: | Cornilescu, G, Cornilescu, C.C, Zhao, Q, Frederick, R.O, Peterson, F.C, Thao, S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a homodimeric hypothetical protein, at5g22580, a structural genomics target from Arabidopsis thaliana.
J.Biomol.Nmr, 29, 2004
|
|
1R8P
 
 | | HPV-16 E2C solution structure | | Descriptor: | Regulatory protein E2 | | Authors: | Nadra, A.D, Eliseo, T, Cicero, D.O. | | Deposit date: | 2003-10-28 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HPV-16 E2 DNA binding domain, a transcriptional regulator with a dimeric beta-barrel fold
J.Biomol.NMR, 30, 2004
|
|
1L3H
 
 | | NMR structure of P41icf, a potent inhibitor of human cathepsin L | | Descriptor: | MHC CLASS II-ASSOCIATED P41 INVARIANT CHAIN FRAGMENT (P41icf) | | Authors: | Chiva, C, Barthe, P, Codina, A, Giralt, E. | | Deposit date: | 2002-02-27 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Synthesis and NMR structure of P41ICF, a potent inhibitor of human cathepsin L
J.Am.Chem.Soc., 125, 2003
|
|
1L3M
 
 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
1KWJ
 
 | | solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans, minimized average structure | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-01-29 | | Release date: | 2002-02-06 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
1GIP
 
 | |
6NAN
 
 | | NMR structure determination of Ixolaris and Factor X interaction reveals a noncanonical mechanism of Kunitz inhibition | | Descriptor: | Ixolaris | | Authors: | De Paula, V.S, Sgourakis, N.G, Francischetti, I.M.B, Almeida, F.C.L, Monteiro, R.Q, Valente, A.P. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of Ixolaris and factor X(a) interaction reveals a noncanonical mechanism of Kunitz inhibition.
Blood, 134, 2019
|
|
1PQT
 
 | |
1GH8
 
 | |
1JE9
 
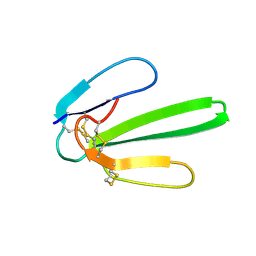 | | NMR SOLUTION STRUCTURE OF NT2 | | Descriptor: | SHORT NEUROTOXIN II | | Authors: | Cheng, Y, Wang, W, Wang, J. | | Deposit date: | 2001-06-16 | | Release date: | 2001-07-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
1FHS
 
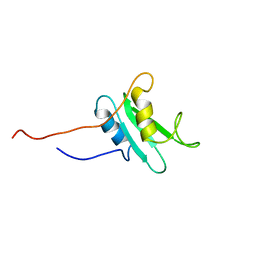 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE SRC HOMOLOGY DOMAIN-2 OF THE GROWTH FACTOR RECEPTOR BOUND PROTEIN-2, NMR, 18 STRUCTURES | | Descriptor: | GROWTH FACTOR RECEPTOR BOUND PROTEIN-2 | | Authors: | Senior, M.M, Frederick, A.F, Black, S, Perkins, L.M, Wilson, O, Snow, M.E, Wang, Y.-S. | | Deposit date: | 1997-06-12 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the Src homology domain-2 of the growth factor receptor-bound protein-2.
J.Biomol.NMR, 11, 1998
|
|
1MZK
 
 | | NMR structure of kinase-interacting FHA domain of kinase associated protein phosphatase, KAPP in Arabidopsis | | Descriptor: | KINASE ASSOCIATED PROTEIN PHOSPHATASE | | Authors: | Lee, G, Ding, Z, Walker, J.C, Van Doren, S.R. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the forkhead-associated domain from the Arabidopsis receptor kinase-associated protein phosphatase.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PUL
 
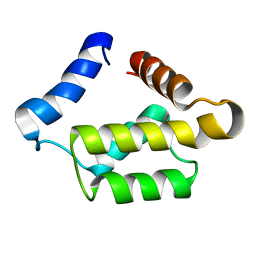 | | Solution structure for the 21KDa caenorhabditis elegans protein CE32E8.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR33 | | Descriptor: | Hypothetical protein C32E8.3 in chromosome I | | Authors: | Tejero, R, Aramini, J.M, Swapna, G.V.T, Monleon, D, Chiang, Y, Macapagal, D, Gunsalus, K.C, Kim, S, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-25 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone 1H, 15N and 13C assignments for the 21 kDa Caenorhabditis elegans homologue of "brain-specific" protein.
J.Biomol.Nmr, 28, 2004
|
|
1JRM
 
 | |
1T0G
 
 | | Hypothetical protein At2g24940.1 from Arabidopsis thaliana has a cytochrome b5 like fold | | Descriptor: | cytochrome b5 domain-containing protein | | Authors: | Song, J, Vinarov, D.A, Tyler, E.M, Shahan, M.N, Tyler, R.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hypothetical protein At2g24940.1 from Arabidopsis thaliana has a cytochrome b5 like fold
J.Biomol.NMR, 30, 2004
|
|
1CP8
 
 | | NMR STRUCTURE OF DNA (5'-D(TTGGCCAA)2-3') COMPLEXED WITH NOVEL ANTITUMOR DRUG UCH9 | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-(SEC-BUTYL)-ANTHRACENE, DNA (5'-D(P*TP*TP*GP*GP*CP*CP*AP*A)-3'), MAGNESIUM ION, ... | | Authors: | Katahira, R, Katahira, M, Yamashita, Y, Ogawa, H, Kyogoku, Y, Yoshida, M. | | Deposit date: | 1999-06-11 | | Release date: | 1999-07-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the novel antitumor drug UCH9 complexed with d(TTGGCCAA)2 as determined by NMR.
Nucleic Acids Res., 26, 1998
|
|
1BWY
 
 | | NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN | | Descriptor: | PROTEIN (HEART FATTY ACID BINDING PROTEIN) | | Authors: | Lassen, D, Luecke, C, Kveder, M, Mesgarzadeh, A, Schmidt, J.M, Specht, B, Lezius, A, Spener, F, Rueterjans, H. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy.
Eur.J.Biochem., 230, 1995
|
|
1ROF
 
 | | NMR STUDY OF 4FE-4S FERREDOXIN OF THERMATOGA MARITIMA | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Roesch, P, Sticht, H, Wildegger, G, Bentrop, D, Darimont, B, Sterner, R. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An NMR-derived model for the solution structure of oxidized Thermotoga maritima 1[Fe4-S4] ferredoxin.
Eur.J.Biochem., 237, 1996
|
|
