7QWS
 
 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
8IFE
 
 | | Arbekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFD
 
 | | Dibekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
4WWA
 
 | | Crystal structure of binary complex Bud32-Cgi121 | | Descriptor: | EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, SULFATE ION | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW7
 
 | |
4WX8
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1 | | Authors: | Zhang, W, Van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WXA
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1, ... | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
7OW7
 
 | |
5BTH
 
 | | Crystal structure of Candida albicans Rai1 | | Descriptor: | Decapping nuclease RAI1 | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
7D63
 
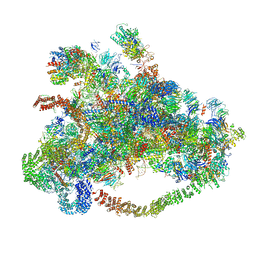 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
4DFX
 
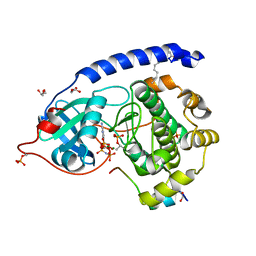 | | Crystal structure of myristoylated K7C catalytic subunit of cAMP-dependent protein kinase in complex with SP20 and AMP-PNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYRISTIC ACID, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Taylor, S.S. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-06 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Role of N-terminal myristylation in the structure and regulation of cAMP-dependent protein kinase.
J.Mol.Biol., 422, 2012
|
|
7D5S
 
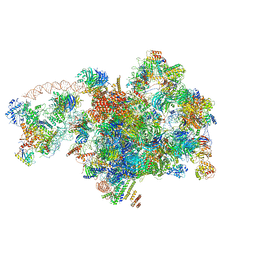 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S12, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2)
To Be Published
|
|
3KZ1
 
 | | Crystal Structure of the Complex of PDZ-RhoGEF DH/PH domains with GTP-gamma-S Activated RhoA | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Rho guanine nucleotide exchange factor 11, ... | | Authors: | Chen, Z, Sternweis, P.C, Sprang, S.R. | | Deposit date: | 2009-12-07 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation.
J.Biol.Chem., 285, 2010
|
|
7TUT
 
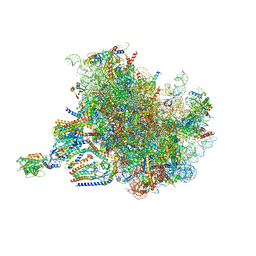 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-02-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
6YBD
 
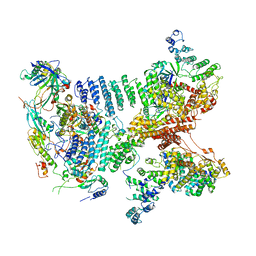 | | Structure of a human 48S translational initiation complex - eIF3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14, 40S ribosomal protein S17, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
7D5T
 
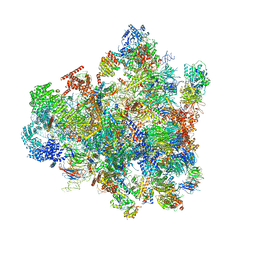 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1)
To Be Published
|
|
8ZWJ
 
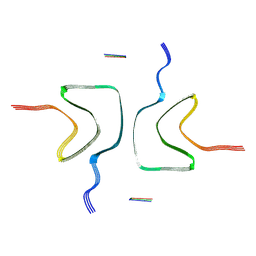 | |
8ZWK
 
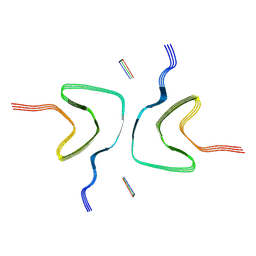 | |
7TM3
 
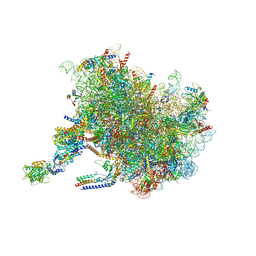 | | Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
1M16
 
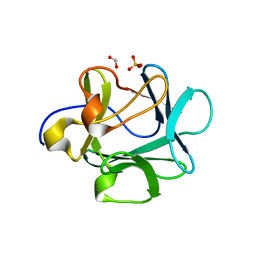 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag and Leu 44 Replaced with Phe (L44F), Leu 73 Replaced with Val (L73V), Val 109 Replaced with Leu (V109L) and Cys 117 Replaced with Val (C117V). | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Kim, J, Spielmann, G.L, Logan, T.M, Blaber, M. | | Deposit date: | 2002-06-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold
Protein Sci., 12, 2003
|
|
4MDE
 
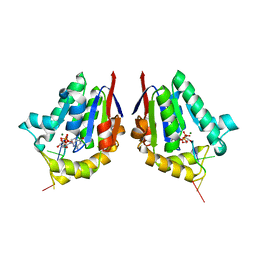 | | Structure of bacterial polynucleotide kinase product complex bound to GDP and DNA | | Descriptor: | DNA (5'-D(P*CP*CP*TP*GP*T)-3'), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shuman, S, Das, U, Wang, L.K, Smith, P, Jacewicz, A. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5'-OH oligonucleotide and a product complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition.
Nucleic Acids Res., 42, 2014
|
|
8UJY
 
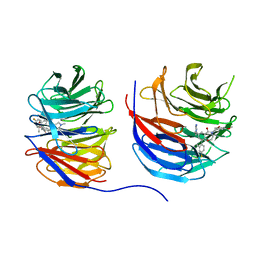 | | Crystal structure of human WD repeat-containing protein 5 in complex with 4-(3,5-dimethoxybenzyl)-9-(4-fluoro-2-methylphenyl)-7-((2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl)methyl)-3,4-dihydrobenzo[f][1,4]oxazepin-5(2H)-one (compound 8) | | Descriptor: | (9P)-4-[(3,5-dimethoxyphenyl)methyl]-9-(4-fluoro-2-methylphenyl)-7-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-3,4-dihydro-1,4-benzoxazepin-5(2H)-one, WD repeat-containing protein 5 | | Authors: | Zhao, B, Amporndanai, K, Fesik, S.W. | | Deposit date: | 2023-10-11 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Discovery of Potent, Orally Bioavailable Benzoxazepinone-Based WD Repeat Domain 5 Inhibitors.
J.Med.Chem., 66, 2023
|
|
4ZUX
 
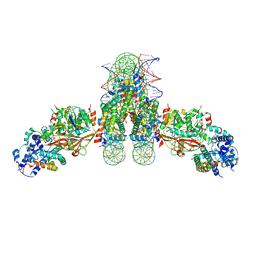 | |
7WTV
 
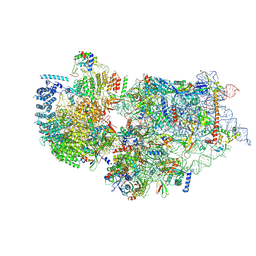 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A2 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
8XXN
 
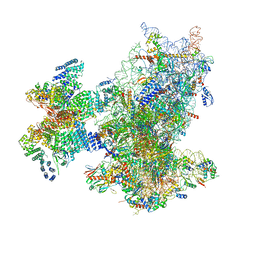 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
