1F2Z
 
 | |
1F30
 
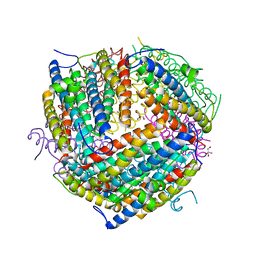 | | THE STRUCTURAL BASIS FOR DNA PROTECTION BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, ZINC ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2000-05-31 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structural Basis for DNA Protection by E. coli Dps Protein
To be Published
|
|
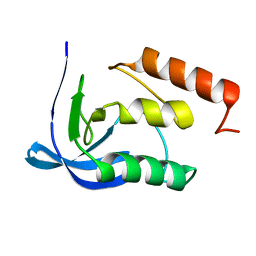
1F31
 
 | |
1F32
 
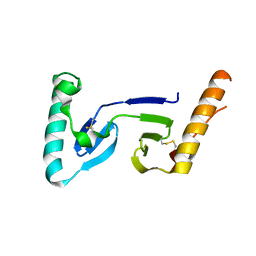 | | CRYSTAL STRUCTURE OF ASCARIS PEPSIN INHIBITOR-3 | | Descriptor: | MAJOR PEPSIN INHIBITOR PI-3 | | Authors: | Ng, K.K, Petersen, J.F, Cherney, M.M, Garen, C, James, M.N. | | Deposit date: | 2000-05-31 | | Release date: | 2001-02-01 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the inhibition of porcine pepsin by Ascaris pepsin inhibitor-3.
Nat.Struct.Biol., 7, 2000
|
|
1F33
 
 | |
1F34
 
 | | CRYSTAL STRUCTURE OF ASCARIS PEPSIN INHIBITOR-3 BOUND TO PORCINE PEPSIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAJOR PEPSIN INHIBITOR PI-3, PEPSIN A | | Authors: | Ng, K.K, Petersen, J.F, Cherney, M.M, Garen, C, James, M.N. | | Deposit date: | 2000-05-31 | | Release date: | 2001-02-01 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the inhibition of porcine pepsin by Ascaris pepsin inhibitor-3.
Nat.Struct.Biol., 7, 2000
|
|
1F35
 
 | |
1F36
 
 | |
1F37
 
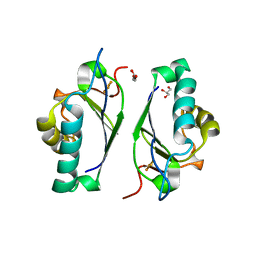 | | STRUCTURE OF A THIOREDOXIN-LIKE [2FE-2S] FERREDOXIN FROM AQUIFEX AEOLICUS | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN [2FE-2S], GLYCEROL | | Authors: | Yeh, A.P, Chatelet, C, Soltis, S.M, Kuhn, P, Meyer, J, Rees, D.C. | | Deposit date: | 2000-05-31 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus.
J.Mol.Biol., 300, 2000
|
|
1F38
 
 | |
1F39
 
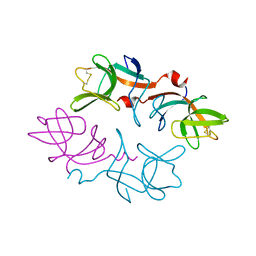 | | CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR C-TERMINAL DOMAIN | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Bell, C.E, Frescura, P, Hochschild, A, Lewis, M. | | Deposit date: | 2000-06-01 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding.
Cell(Cambridge,Mass.), 101, 2000
|
|
1F3A
 
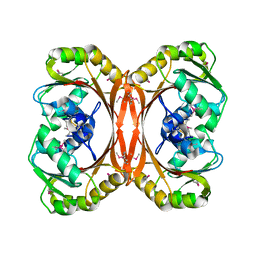
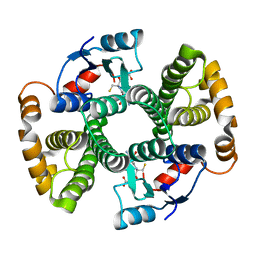 | | CRYSTAL STRUCTURE OF MGSTA1-1 IN COMPLEX WITH GSH | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE YA CHAIN | | Authors: | Gu, Y, Singh, S.V, Ji, X. | | Deposit date: | 2000-06-01 | | Release date: | 2000-10-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Residue R216 and catalytic efficiency of a murine class alpha glutathione S-transferase toward benzo[a]pyrene 7(R),8(S)-diol 9(S), 10(R)-epoxide.
Biochemistry, 39, 2000
|
|
1F3B
 
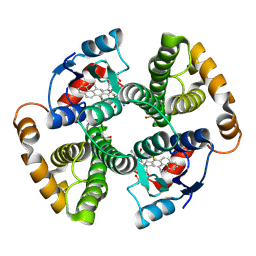 | | CRYSTAL STRUCTURE OF MGSTA1-1 IN COMPLEX WITH GLUTATHIONE CONJUGATE OF BENZO[A]PYRENE EPOXIDE | | Descriptor: | 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, GLUTATHIONE S-TRANSFERASE YA CHAIN | | Authors: | Gu, Y, Singh, S.V, Ji, X. | | Deposit date: | 2000-06-01 | | Release date: | 2000-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Residue R216 and catalytic efficiency of a murine class alpha glutathione S-transferase toward benzo[a]pyrene 7(R),8(S)-diol 9(S), 10(R)-epoxide.
Biochemistry, 39, 2000
|
|
1F3C
 
 | |
1F3D
 
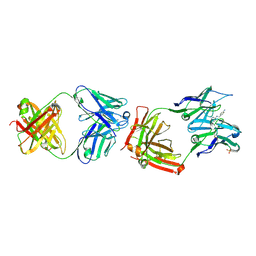 | | CATALYTIC ANTIBODY 4B2 IN COMPLEX WITH ITS AMIDINIUM HAPTEN. | | Descriptor: | 2-(4-AMINOBENZYLAMINO)-3,4,5,6-TETRAHYDROPYRIDINIUM, CATALYTIC ANTIBODY 4B2, SULFATE ION | | Authors: | Golinelli-Pimpaneau, B, Goncalves, O, Dintinger, T, Blanchard, D, Knossow, M, Tellier, C. | | Deposit date: | 2000-06-02 | | Release date: | 2000-09-13 | | Last modified: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural evidence for a programmed general base in the active site of a catalytic antibody.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F3E
 
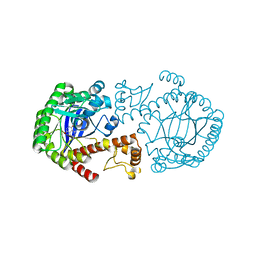 | | A NEW TARGET FOR SHIGELLOSIS: RATIONAL DESIGN AND CRYSTALLOGRAPHIC STUDIES OF INHIBITORS OF TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | 3,5-DIAMINOPHTHALHYDRAZIDE, QUEUINE TRNA-RIBOSYLTRANSFERASE, ZINC ION | | Authors: | Graedler, U, Gerber, H.-D, Goodenough-Lashua, D.M, Garcia, G.A.G, Ficner, R, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2000-06-02 | | Release date: | 2000-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase.
J.Mol.Biol., 306, 2001
|
|
1F3F
 
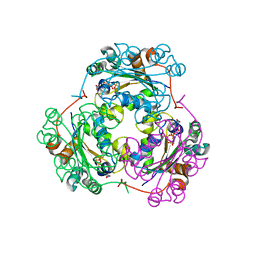 | | STRUCTURE OF THE H122G NUCLEOSIDE DIPHOSPHATE KINASE / D4T-TRIPHOSPHATE.MG COMPLEX | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-DIPHOSPHATE, 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meyer, P, Schneider, B, Sarfati, S, Deville-Bonne, D, Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B. | | Deposit date: | 2000-06-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
1F3G
 
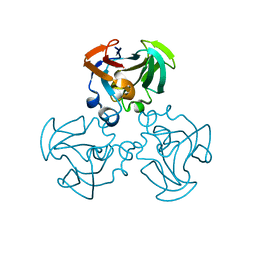 | | THREE-DIMENSIONAL STRUCTURE OF THE ESCHERICHIA COLI PHOSPHOCARRIER PROTEIN III GLC | | Descriptor: | GLUCOSE-SPECIFIC PHOSPHOCARRIER PROTEIN IIAGLC | | Authors: | Worthylake, D, Meadow, N, Roseman, S, Liao, D.-I, Herzberg, O, Remington, S.J. | | Deposit date: | 1991-08-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the Escherichia coli phosphocarrier protein IIIglc.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1F3H
 
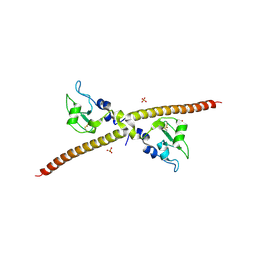 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN ANTI-APOPTOTIC PROTEIN SURVIVIN | | Descriptor: | SULFATE ION, SURVIVIN, ZINC ION | | Authors: | Verdecia, M.A, Huang, H, Dutil, E, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-03 | | Release date: | 2000-12-06 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement.
Nat.Struct.Biol., 7, 2000
|
|
1F3J
 
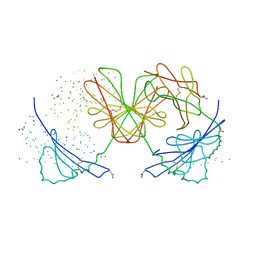 | | HISTOCOMPATIBILITY ANTIGEN I-AG7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, LYSOZYME C, ... | | Authors: | Latek, R.R, Unanue, E.R, Fremont, D.H. | | Deposit date: | 2000-06-04 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice.
Immunity, 12, 2000
|
|
1F3K
 
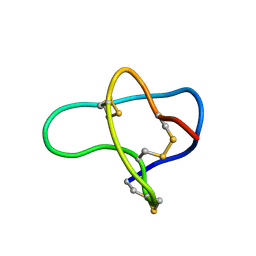 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF OMEGA-CONOTOXIN TXVII, AN L-TYPE CALCIUM CHANNEL BLOCKER | | Descriptor: | OMEGA-CONOTOXIN TXVII | | Authors: | Kobayashi, K, Sasaki, T, Sato, K, Kohno, T. | | Deposit date: | 2000-06-05 | | Release date: | 2000-12-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of omega-conotoxin TxVII, an L-type calcium channel blocker.
Biochemistry, 39, 2000
|
|
1F3L
 
 | |
1F3M
 
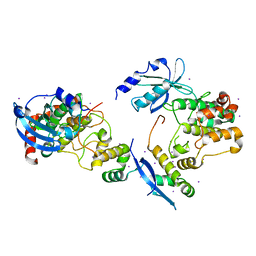 | | CRYSTAL STRUCTURE OF HUMAN SERINE/THREONINE KINASE PAK1 | | Descriptor: | IODIDE ION, SERINE/THREONINE-PROTEIN KINASE PAK-ALPHA | | Authors: | Lei, M, Lu, W, Meng, W, Parrini, M.-C, Eck, M.J, Mayer, B.J, Harrison, S.C. | | Deposit date: | 2000-06-05 | | Release date: | 2000-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch.
Cell(Cambridge,Mass.), 102, 2000
|
|
1F3O
 
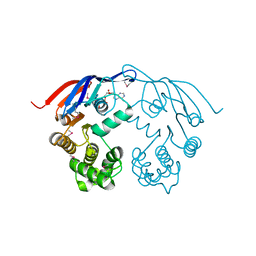 | | Crystal structure of MJ0796 ATP-binding cassette | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYPOTHETICAL ABC TRANSPORTER ATP-BINDING PROTEIN MJ0796, MAGNESIUM ION | | Authors: | Yuan, Y.-R, Hunt, J.F. | | Deposit date: | 2000-06-05 | | Release date: | 2001-07-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the MJ0796 ATP-binding cassette. Implications for the structural consequences of ATP hydrolysis in the active site of an ABC transporter.
J.Biol.Chem., 276, 2001
|
|
1F3P
 
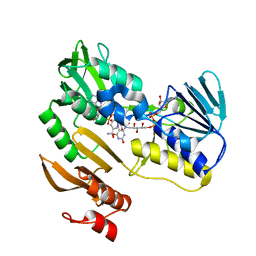 | | FERREDOXIN REDUCTASE (BPHA4)-NADH COMPLEX | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsuidagger, Y. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
