6K6T
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-IMP bound form | | Descriptor: | 9-[(1R,6R,8R,9S,10R,15S,17R,18S)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-17-(6-oxidanylidene-3H-purin-9-yl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]-3H-purin-6-one, CALCIUM ION, EAL domain protein | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-IMP bound form
To Be Published
|
|
4GU5
 
 | | Structure of Full-length Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zoltowski, B.D, Vaidya, A.T, Top, D, Widom, J, Young, M.W, Levy, C, Jones, A.R, Scrutton, N.S, Leys, D, Crane, B.R. | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Updated structure of Drosophila cryptochrome.
Nature, 495, 2013
|
|
6JIE
 
 | | YaeO bound to Magnesium from Vibrio cholerae O395 | | Descriptor: | MAGNESIUM ION, YaeO | | Authors: | Pal, K, Yadav, M, Sen, U. | | Deposit date: | 2019-02-20 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Vibrio cholerae YaeO is a Structural Homologue of RNA Chaperone Hfq that Inhibits Rho-dependent Transcription Termination by Dissociating its Hexameric State.
J.Mol.Biol., 431, 2019
|
|
5DTB
 
 | |
4X23
 
 | |
6XYH
 
 | | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2) | | Descriptor: | AMS3 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2)
To Be Published
|
|
3M9Q
 
 | |
4XCW
 
 | |
4XDA
 
 | | Vibrio cholerae O395 Ribokinase complexed with Ribose, ADP and Sodium ion. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribokinase, SODIUM ION, ... | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Apo and Ligand Bound Vibrio cholerae Ribokinase (Vc-RK): Role of Monovalent Cation Induced Activation and Structural Flexibility in Sugar Phosphorylation
Adv.Exp.Med.Biol., 842, 2015
|
|
2M48
 
 | | Solution Structure of IBR-RING2 Tandem Domain from Parkin | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE PARKIN, ZINC ION | | Authors: | Noh, Y.J, Mercier, P, Spratt, D.E, Shaw, G.S. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A molecular explanation for the recessive nature of parkin-linked Parkinson's disease.
Nat Commun, 4, 2013
|
|
3MN6
 
 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3EL2
 
 | | Crystal Structure of Monomeric Actin Bound to Ca-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
2LWR
 
 | | Solution Structure of RING2 Domain from Parkin | | Descriptor: | SD01679p, ZINC ION | | Authors: | Mercier, P, Spratt, D.E, Manczyk, N, Shaw, G.S. | | Deposit date: | 2012-08-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A molecular explanation for the recessive nature of parkin-linked Parkinson's disease.
Nat Commun, 4, 2013
|
|
3EIN
 
 | | Delta class GST | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1-1 | | Authors: | Feil, S.C. | | Deposit date: | 2008-09-17 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.126 Å) | | Cite: | Probing insect detoxification systems
To be Published
|
|
5X9X
 
 | |
3EKU
 
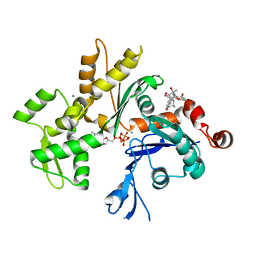 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
6X7H
 
 | |
2MEF
 
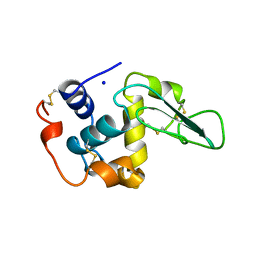 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-04 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
4XCK
 
 | | Vibrio cholerae O395 Ribokinase complexed with ADP, Ribose and Cesium ion. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CESIUM ION, Ribokinase, ... | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structure of Apo and Ligand Bound Vibrio cholerae Ribokinase (Vc-RK): Role of Monovalent Cation Induced Activation and Structural Flexibility in Sugar Phosphorylation.
Adv. Exp. Med. Biol., 842, 2015
|
|
4XHV
 
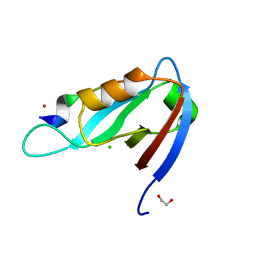 | | Crystal structure of Drosophila Spinophilin-PDZ and a C-terminal peptide of Neurexin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LP20995p, ... | | Authors: | Driller, J.H, Muhammad, K.G.H, Reddy, S, Rey, U, Boehme, M.A, Hollmann, C, Ramesh, N, Depner, H, Luetzkendorf, J, Matkovic, T, Bergeron, D, Quentin, C, Schmoranzer, J, Goettfert, F, Holt, M, Wahl, M.C, Hell, S.W, Walter, A, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function.
Nat Commun, 6, 2015
|
|
3EAD
 
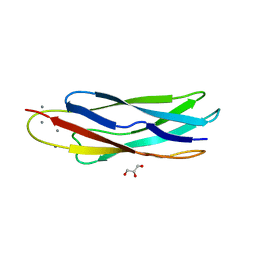 | | Crystal structure of CALX-CBD1 | | Descriptor: | CALCIUM ION, GLYCEROL, Na/Ca exchange protein | | Authors: | Zheng, L, Wang, M. | | Deposit date: | 2008-08-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
2MEG
 
 | | CHANGES IN CONFORMATIONAL STABILITY OF A SERIES OF MUTANT HUMAN LYSOZYMES AT CONSTANT POSITIONS. | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
5EX7
 
 | | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA | | Descriptor: | Brain tumor protein, RNA (5'-R(P*UP*UP*UP*GP*UP*UP*GP*U)-3') | | Authors: | Wang, Y, Yu, Z, Wang, M, Liu, C.P, Yang, N, Xu, R.M. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA
To Be Published
|
|
2MEE
 
 | |
5F11
 
 | |
