2QSJ
 
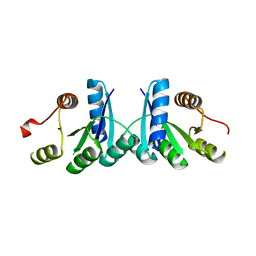 | | Crystal structure of a LuxR family DNA-binding response regulator from Silicibacter pomeroyi | | Descriptor: | DNA-binding response regulator, LuxR family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a LuxR family DNA-binding response regulator from Silicibacter pomeroyi.
To be Published
|
|
8CMN
 
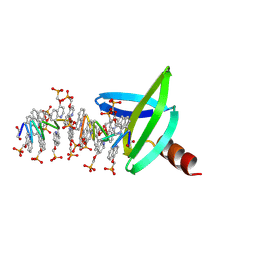 | |
2Y3P
 
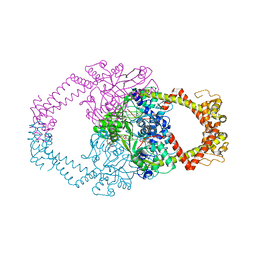 | | Crystal structure of N-terminal domain of GyrA with the antibiotic simocyclinone D8 | | Descriptor: | DNA GYRASE SUBUNIT A, MAGNESIUM ION, SIMOCYCLINONE D8 | | Authors: | Edwards, M.J, Flatman, R.H, Mitchenall, L.A, Stevenson, C.E.M, Le, T.B.K, Clarke, T.A, McKay, A.R, Fiedler, H.-P, Buttner, M.J, Lawson, D.M, Maxwell, A. | | Deposit date: | 2010-12-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8, Bound to DNA Gyrase.
Science, 326, 2009
|
|
2MAX
 
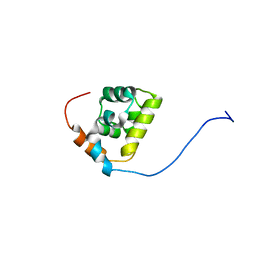 | |
5U91
 
 | | Crystal structure of Tre/loxLTR complex | | Descriptor: | DNA (37-MER), Tre recombinase protein | | Authors: | Meinke, G, Karpinski, J, Buchholz, F, Bohm, A. | | Deposit date: | 2016-12-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Crystal structure of an engineered, HIV-specific recombinase for removal of integrated proviral DNA.
Nucleic Acids Res., 45, 2017
|
|
6F3C
 
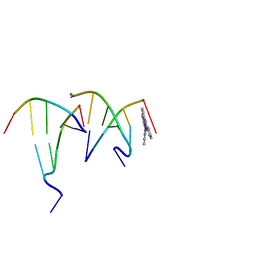 | | The cytotoxic [Pt(H2bapbpy)] platinum complex interacting with the CGTACG hexamer | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, [Pt(H2bapbpy)] platinum | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Papi, F. | | Deposit date: | 2017-11-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Induction of a Four-Way Junction Structure in the DNA Palindromic Hexanucleotide 5'-d(CGTACG)-3' by a Mononuclear Platinum Complex.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3P9A
 
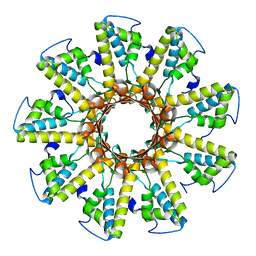 | |
423D
 
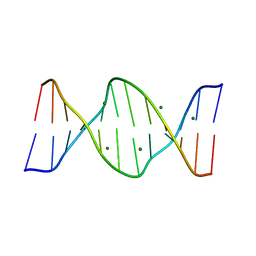 | | 5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3' | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3'), MAGNESIUM ION | | Authors: | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | Deposit date: | 1998-09-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8C84
 
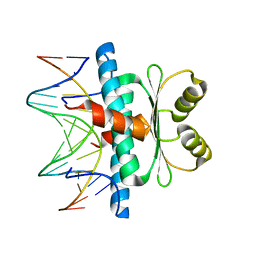 | | Crystal structure of MADS-box/MEF2D N-terminal domain complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), MEF2D protein | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
4MQD
 
 | | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis | | Descriptor: | DNA-entry nuclease inhibitor | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis
To be Published
|
|
4JOM
 
 | | Structure of E. coli Pol III 3mPHP mutant | | Descriptor: | DNA polymerase III subunit alpha, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Barros, T, Guenther, J, Kelch, B, Anaya, J, Prabhakar, A, O'Donnell, M, Kuriyan, J, Lamers, M.H. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structural role for the PHP domain in E. coli DNA polymerase III.
Bmc Struct.Biol., 13, 2013
|
|
6IMK
 
 | | The crystal structure of AsfvLIG:CG complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*G)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
1HM1
 
 | | THE SOLUTION NMR STRUCTURE OF A THERMALLY STABLE FAPY ADDUCT OF AFLATOXIN B1 IN AN OLIGODEOXYNUCLEOTIDE DUPLEX REFINED FROM DISTANCE RESTRAINED MOLECULAR DYNAMICS SIMULATED ANNEALING, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*TP*AP*TP*(FAG)P*AP*TP*TP*CP*A)-3'), DNA (5'-D(TP*GP*AP*AP*TP*CP*AP*TP*AP*G)-3') | | Authors: | Mao, H, Deng, Z, Wang, F, Harris, T.M, Stone, M.P. | | Deposit date: | 1998-05-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR.
Biochemistry, 37, 1998
|
|
6IML
 
 | | The crystal structure of AsfvLIG:CT1 complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
4O9I
 
 | |
7ZQN
 
 | |
7ZQO
 
 | |
2QRV
 
 | | Structure of Dnmt3a-Dnmt3L C-terminal domain complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jia, D, Cheng, X. | | Deposit date: | 2007-07-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Nature, 449, 2007
|
|
5H46
 
 | | Mycobacterium smegmatis Dps1 mutant - F47E | | Descriptor: | DNA protection during starvation protein, FE (II) ION | | Authors: | Williams, S.M, Chandran, A.V, Vijayan, M, Chatterji, D. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Mutation Directs the Structural Switch of DNA Binding Proteins under Starvation to a Ferritin-like Protein Cage.
Structure, 25, 2017
|
|
2YW6
 
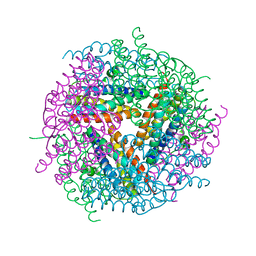 | | Structural studies of N terminal deletion mutant of Dps from Mycobacterium smegmatis | | Descriptor: | DNA protection during starvation protein | | Authors: | Roy, S, Saraswathi, R, Gupta, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2007-04-19 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Role of N and C-terminal Tails in DNA Binding and Assembly in Dps: Structural Studies of Mycobacterium smegmatis Dps Deletion Mutants
J.Mol.Biol., 370, 2007
|
|
4LG7
 
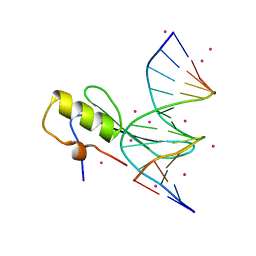 | | Crystal structure MBD4 MBD domain in complex with methylated CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 4, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure MBD4 MBD domain in complex with methylated CpG DNA
To be Published
|
|
8B0A
 
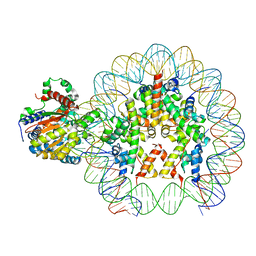 | | Cryo-EM structure of ALC1 bound to an asymmetric, site-specifically PARylated nucleosome | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (149-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Asymmetric nucleosome PARylation at DNA breaks mediates directional nucleosome sliding by ALC1.
Nat Commun, 15, 2024
|
|
7XVL
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment) | | Descriptor: | DNA (169-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment)
To Be Published
|
|
4ID3
 
 | |
1CKQ
 
 | |
