3TU9
 
 | |
3L8F
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli complexed with magnesium and phosphate | | Descriptor: | D,D-heptose 1,7-bisphosphate phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
1ML6
 
 | | Crystal Structure of mGSTA2-2 in Complex with the Glutathione Conjugate of Benzo[a]pyrene-7(R),8(S)-Diol-9(S),10(R)-Epoxide | | Descriptor: | 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, Glutathione S-Transferase GT41A, ISOPROPYL ALCOHOL | | Authors: | Gu, Y, Xiao, B, Wargo, H.L, Bucher, M.H, Singh, S.V, Ji, X. | | Deposit date: | 2002-08-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Residues 207, 216, and 221 and the catalytic activity of mGSTA1-1 and mGSTA2-2 toward
benzo[a]pyrene-(7R,8S)-diol-(9S,10R)-epoxide
Biochemistry, 42, 2003
|
|
1ISP
 
 | | Crystal structure of Bacillus subtilis lipase at 1.3A resolution | | Descriptor: | GLYCEROL, lipase | | Authors: | Kawasaki, K, Kondo, H, Suzuki, M, Ohgiya, S, Tsuda, S. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3UCG
 
 | |
3LLC
 
 | |
3LKH
 
 | |
4JO2
 
 | |
3QIH
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | Descriptor: | (4aS,7aS)-1,4-bis(3-hydroxybenzyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease, ... | | Authors: | Lindemann, I, Heine, A, Klebe, G. | | Deposit date: | 2011-01-27 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Novel inhibitors for HIV-1 protease
To be Published
|
|
3EZS
 
 | |
3QEA
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with DNA (complex II) | | Descriptor: | BARIUM ION, DNA (5'-D(P*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
3LIT
 
 | | The crystal structure of htlv protease complexed with the inhibitor KNI-10681 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-[(2R)-3-methylbutan-2-yl]-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3QN8
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | Descriptor: | (4aS,7aS)-1,4-bis(3-hydroxybenzyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease | | Authors: | Lindemann, I, Heine, A, Klebe, G. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Novel inhibitors for HIV-1 protease
To be Published
|
|
1QIW
 
 | | Calmodulin complexed with N-(3,3,-diphenylpropyl)-N'-[1-R-(3,4-bis-butoxyphenyl)-ethyl]-propylenediamine (DPD) | | Descriptor: | CALCIUM ION, CALMODULIN, N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(2 3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE | | Authors: | Harmat, V, Bocskei, Z.S, Vertessy, B.G, Ovadi, J, Naray-Szabo, G. | | Deposit date: | 1999-06-17 | | Release date: | 2000-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Potent Calmodulin Antagonist with Arylalkylamine Structure: Crystallographic, Spectroscopic and Functional Studies
J.Mol.Biol., 297, 2000
|
|
1N4K
 
 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Bosanac, I, Alattia, J.R, Mal, T.K, Chan, J, Talarico, S, Tong, F.K, Tong, K.I, Yoshikawa, F, Furuichi, T, Iwai, M, Michikawa, T, Mikoshiba, K, Ikura, M. | | Deposit date: | 2002-10-31 | | Release date: | 2002-12-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the inositol 1,4,5-trisphosphate receptor
binding core in complex with its ligand.
Nature, 420, 2002
|
|
4JCO
 
 | |
4JDE
 
 | | Crystal structure of PUD-1/PUD-2 heterodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein F15E11.1, ... | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2013-02-25 | | Release date: | 2013-11-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of PUD-1 and PUD-2, two proteins up-regulated in a long-lived daf-2 mutant.
Plos One, 8, 2013
|
|
4CPT
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl [(2S)-1-(2-{3-[(3R,6S,10Z)-3-hydroxy-4,7-dioxo-6-(propan-2-yl)-5,8-diazabicyclo[11.2.2]heptadeca-1(15),10,13,16-tetraen-3-yl]propyl}-2-[4-(pyridin-3-yl)benzyl]hydrazinyl)-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
1VRU
 
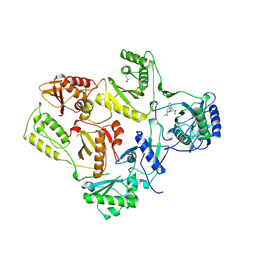 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
2A98
 
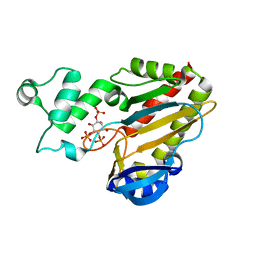 | | Crystal structure of the catalytic domain of human inositol 1,4,5-trisphosphate 3-kinase C | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate 3-kinase C | | Authors: | Hallberg, B.M, Ogg, D, Ehn, M, Graslund, S, Hammarstrom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Persson, C, Sagemark, J, Schuler, H, Stenmark, P, Thorsell, A.-G, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the catalytic domain of human inositol 1,4,5-trisphosphate 3-kinase C
To be Published
|
|
6KHI
 
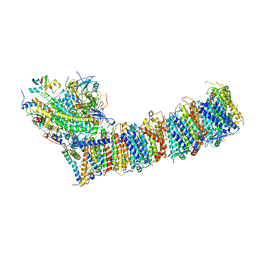 | | Supercomplex for cylic electron transport in cyanobacteria | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
3LUR
 
 | |
3EZ0
 
 | |
1H8V
 
 | | The X-ray Crystal Structure of the Trichoderma reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Shaw, A, Ropp, T.H, Wu, S, Bott, R, Cameron, A.D, Stahlberg, J, Mitchinson, C, Jones, T.A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution
J.Mol.Biol., 308, 2001
|
|
2AEB
 
 | | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in immune response. | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
