4KT0
 
 | | Crystal structure of a virus like photosystem I from the cyanobacterium Synechocystis PCC 6803 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Mazor, Y, Nataf, D, Toporik, H, Nelson, N. | | Deposit date: | 2013-05-19 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of virus-like photosystem I complexes from the mesophilic cyanobacterium Synechocystis PCC 6803.
Elife, 3, 2014
|
|
4IXR
 
 | | RT fs X-ray diffraction of Photosystem II, first illuminated state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
4IXQ
 
 | | RT fs X-ray diffraction of Photosystem II, dark state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.7 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
1Z69
 
 | | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Ermler, U, Hagemeier, C.H, Thauer, R.K, Shima, S. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: Architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family
Protein Sci., 14, 2005
|
|
4K95
 
 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
2A69
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A68
 
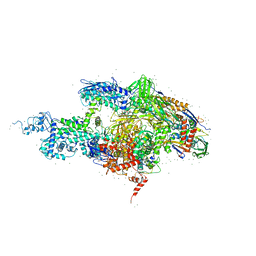 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
4KHP
 
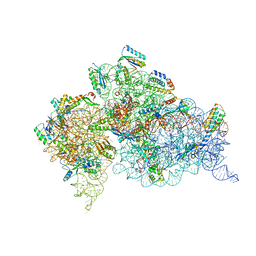 | | Structure of the Thermus thermophilus 30S ribosomal subunit in complex with de-6-MSA-pactamycin | | Descriptor: | 16S Ribosomal RNA, 30S Ribosomal protein S10, 30S Ribosomal protein S11, ... | | Authors: | Tourigny, D.S, Fernandez, I.S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2013-05-01 | | Release date: | 2013-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Bioactive Pactamycin Analog Bound to the 30S Ribosomal Subunit.
J.Mol.Biol., 425, 2013
|
|
4KVB
 
 | |
2AXT
 
 | | Crystal Structure of Photosystem II from Thermosynechococcus elongatus | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Loll, B, Kern, J, Saenger, W, Zouni, A, Biesiadka, J. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-27 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II
NATURE, 438, 2005
|
|
5OY0
 
 | | Structure of synechocystis photosystem I trimer at 2.5A resolution | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nelson, N, Malavath, T, Caspy, I. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis.
Biochim. Biophys. Acta, 1859, 2018
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW7
 
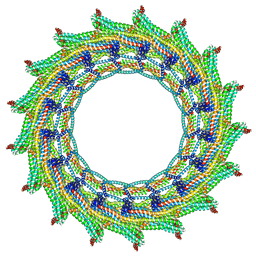 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
5IT9
 
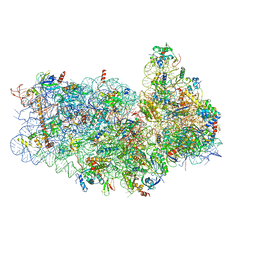 | | Structure of the yeast Kluyveromyces lactis small ribosomal subunit in complex with the cricket paralysis virus IRES. | | Descriptor: | 18S ribosomal RNA, Cricket paralysis virus IRES RNA, MAGNESIUM ION, ... | | Authors: | Murray, J, Savva, C.G, Shin, B.S, Dever, T.E, Ramakrishnan, V, Fernandez, I.S. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural characterization of ribosome recruitment and translocation by type IV IRES.
Elife, 5, 2016
|
|
5IT8
 
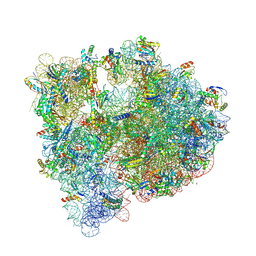 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IT7
 
 | | Structure of the Kluyveromyces lactis 80S ribosome in complex with the cricket paralysis virus IRES and eEF2 | | Descriptor: | (2S)-1-amino-N,N,N-trimethyl-1-oxobutan-2-aminium, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Murray, J, Savva, C.G, Shin, B.S, Dever, T.E, Ramakrishnan, V, Fernandez, I.S. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural characterization of ribosome recruitment and translocation by type IV IRES.
Elife, 5, 2016
|
|
5J4Z
 
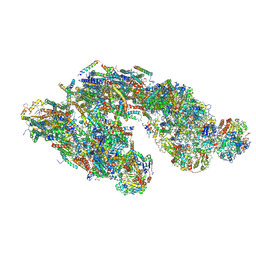 | | Architecture of tight respirasome | | Descriptor: | COMPLEX I 13KDA/NDUFS6, COMPLEX I 15KDA/NDUFS5, COMPLEX I 18KDA/NDUFS6, ... | | Authors: | Letts, J.A, Fiedorczuk, K, Sazanov, L.A. | | Deposit date: | 2016-04-01 | | Release date: | 2016-09-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The architecture of respiratory supercomplexes.
Nature, 537, 2016
|
|
5J7Y
 
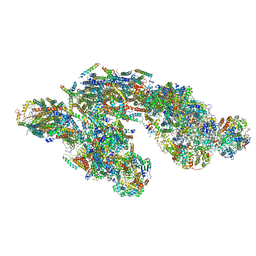 | | Architecture of loose respirasome | | Descriptor: | COMPLEX I 13KDA/NDUFS6, COMPLEX I 15KDA/NDUFS5, COMPLEX I 18KDA/NDUFS6, ... | | Authors: | Letts, J.A, Fiedorczuk, F, Sazanov, L.A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-09-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The architecture of respiratory supercomplexes.
Nature, 537, 2016
|
|
5IV5
 
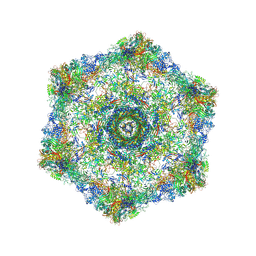 | | Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex | | Descriptor: | Baseplate hub protein gp27, Baseplate tail-tube protein gp48, Baseplate tail-tube protein gp54, ... | | Authors: | Taylor, N.M.I, Guerrero-Ferreira, R.C, Goldie, K.N, Stahlberg, H, Leiman, P.G. | | Deposit date: | 2016-03-19 | | Release date: | 2016-05-18 | | Last modified: | 2018-02-07 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the T4 baseplate and its function in triggering sheath contraction.
Nature, 533, 2016
|
|
5J88
 
 | | Structure of the E coli 70S ribosome with the U1060A mutation in 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-06 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J8B
 
 | | Crystal structure of Elongation Factor 4 (EF-4/LepA) in complex with GDPCP bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Lin, J, Steitz, T.A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Elongation factor 4 remodels the A-site tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J91
 
 | | Structure of the Wild-type 70S E coli ribosome bound to Tigecycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-07-06 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
5J5B
 
 | | Structure of the WT E coli ribosome bound to tetracycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-27 | | Last modified: | 2018-08-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5OBM
 
 | | Crystal structure of Gentamicin bound to the yeast 80S ribosome | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-06-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
