8WDY
 
 | | SARS-CoV-2 Omicron BQ.1.1 RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
5C9P
 
 | | Crystal structure of recombinant PLL lectin complexed with L-fucose from Photorhabdus luminescens at 1.75 A resolution | | Descriptor: | GLYCEROL, PLL lectin, alpha-L-fucopyranose | | Authors: | Kumar, A, Sykorova, P, Demo, G, Dobes, P, Hyrsl, P, Wimmerova, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed beta-Propeller Tetrameric Structure.
J.Biol.Chem., 291, 2016
|
|
3LIW
 
 | | Factor XA in complex with (R)-2-(1-ADAMANTYLCARBAMOYLAMINO)-3-(3-CARBAMIDOYL-PHENYL)-N-PHENETHYL-PROPIONIC ACID AMIDE | | Descriptor: | (R)-2-(3-ADAMANTAN-1-YL-UREIDO)-3-(3-CARBAMIMIDOYL-PHENYL)-N-PHENETHYL-PROPIONAMIDE, Activated factor Xa heavy chain, CALCIUM ION, ... | | Authors: | Mueller, M.M, Sperl, S, Sturzebecher, J, Bode, W, Moroder, L. | | Deposit date: | 2010-01-25 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | (R)-3-Amidinophenylalanine-Derived Inhibitors of Factor Xa with a Novel Active-Site Binding Mode
Biol.Chem., 383, 2003
|
|
9CT8
 
 | | Tricomplex of Compound 2, KRAS G12D, and CypA | | Descriptor: | (2R)-2-amino-N-(2-{[(1R)-1-cyclopentyl-2-{[(1M,8S,10S,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-2-oxoethyl](methyl)amino}-2-oxoethyl)-N-methylpropanamide (non-preferred name), CHLORIDE ION, Isoform 2B of GTPase KRas, ... | | Authors: | Zhang, D, Bar Ziv, T, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A neomorphic protein interface catalyzes covalent inhibition of RAS G12D aspartic acid in tumors.
Science, 389, 2025
|
|
5SIS
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(n(nc1)C)C(Nc3cc2nc(cn2cc3)c4ccccc4)=O)C(=O)N(C)CCNC, micromolar IC50=0.002096 | | Descriptor: | MAGNESIUM ION, N~4~,1-dimethyl-N~4~-[2-(methylamino)ethyl]-N~5~-[(4R)-2-phenylimidazo[1,2-a]pyridin-7-yl]-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4MZM
 
 | | MazF from S. aureus crystal form I, P212121, 2.1 A | | Descriptor: | mRNA interferase MazF | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
9CTA
 
 | | Tricomplex of RMC-9945, KRAS G12D, and CypA | | Descriptor: | (2R)-2-{(5S)-7-[(2R)-3-cyclopropyl-2-(methylamino)propanoyl]-2,7-diazaspiro[4.4]nonan-2-yl}-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1R)-1-methoxyethyl]pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-21-(2,2,2-trifluoroethyl)-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-methylbutanamide (non-preferred name), Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, D, Saldajeno-Concar, M, Tomlinson, A.C.A, Yano, J.K, Knox, J.E. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | A neomorphic protein interface catalyzes covalent inhibition of RAS G12D aspartic acid in tumors.
Science, 389, 2025
|
|
5SJU
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c(nc(s1)NC(c2c(ncc(n2)C3CC3)Nc4cncnc4)=O)CN5CCCCC5, micromolar IC50=0.03208 | | Descriptor: | 6-cyclopropyl-N-{4-[(piperidin-1-yl)methyl]-1,3-thiazol-2-yl}-3-[(pyrimidin-5-yl)amino]pyrazine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SK3
 
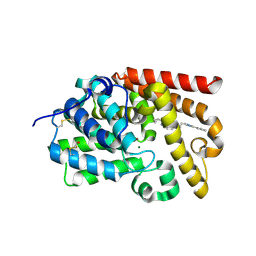 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(cc(c1ccccc1)nc2CCc5nn3c(nc(c3CO)C4CC4)cc5)C, micromolar IC50=0.655585 | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4I26
 
 | | 2.20 Angstroms X-ray crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, 2-aminomuconate 6-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Davis, I, Huo, L, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
5SE9
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1cc(nn2c1nc(c2C)C)OCc3nc(cn3C)c4ccccc4, micromolar IC50=0.007179 | | Descriptor: | (4R)-2,3-dimethyl-6-[(1-methyl-4-phenyl-1H-imidazol-2-yl)methoxy]imidazo[1,2-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
4ESY
 
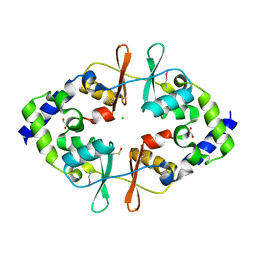 | | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CBS domain containing membrane protein, CHLORIDE ION | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus
To be Published
|
|
9CT9
 
 | | Tricomplex of Compound 3, KRAS G12D, and CypA | | Descriptor: | (2R)-2-{(5S)-7-[(2R)-2-aminopropanoyl]-1-oxo-2,7-diazaspiro[4.4]nonan-2-yl}-2-cyclopentyl-N-[(1M,8S,10S,14R,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]acetamide (non-preferred name), CHLORIDE ION, Isoform 2B of GTPase KRas, ... | | Authors: | Zhang, D, Bar Ziv, T, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A neomorphic protein interface catalyzes covalent inhibition of RAS G12D aspartic acid in tumors.
Science, 389, 2025
|
|
3HSM
 
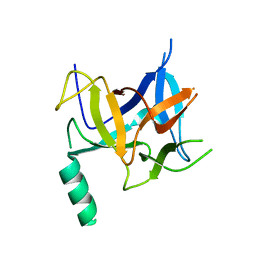 | | Crystal structure of distal N-terminal beta-trefoil domain of Ryanodine Receptor type 1 | | Descriptor: | Ryanodine receptor 1 | | Authors: | Amador, F.J, Liu, S, Ishiyama, N, Plevin, M.J, Wilson, A, MacLennan, D.H, Ikura, M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of type I ryanodine receptor amino-terminal beta-trefoil domain reveals a disease-associated mutation "hot spot" loop
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5CF5
 
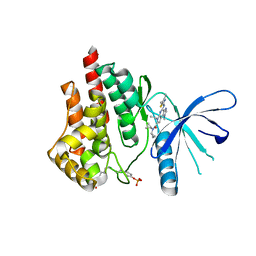 | | CRYSTAL STRUCTURE OF JANUS KINASE 2 IN COMPLEX WITH N,N-DICYCLOPROPYL-7-[(DIMETHYL-1,3-THIAZOL-2-YL)AMINO]-10-ETHYL-3-METHYL-3,5,8,10-TETRAAZATRICYCLO[7.3.0.02,6] DODECA-1(9),2(6),4,7,11-PENTAENE-11-CARBOXAMIDE | | Descriptor: | N,N-dicyclopropyl-4-[(4,5-dimethyl-1,3-thiazol-2-yl)amino]-6-ethyl-1-methyl-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
7E2T
 
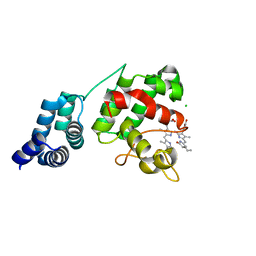 | | Synechocystis GUN4 in complex with phycocyanobilin | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
3LLM
 
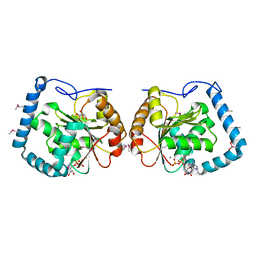 | | Crystal Structure Analysis of a RNA Helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, CACODYLATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-box variant protein.
J.Mol.Biol., 400, 2010
|
|
3P7P
 
 | |
5PWQ
 
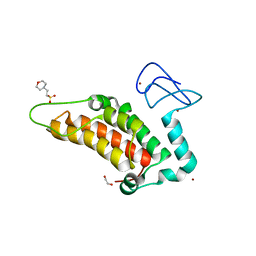 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 14) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5SHI
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(ccc(c2nc(c(n12)C)C)CCc3nc(cn3C)c4ccccc4)C, micromolar IC50=0.954878 | | Descriptor: | (4R)-2,3,5-trimethyl-6-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl]imidazo[1,2-a]pyridine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3HPH
 
 | | Closed tetramer of Visna virus integrase (residues 1-219) in complex with LEDGF IBD | | Descriptor: | GLYCEROL, Integrase, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Wang, J, Cherepanov, P. | | Deposit date: | 2009-06-04 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for functional tetramerization of lentiviral integrase
Plos Pathog., 5, 2009
|
|
3WZ6
 
 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (5) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuhnert, M, Steuber, H, Diederich, W.E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5SJC
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(c(nn1C)C)Cl)C(Nc3nc2nc(cn2cc3)c4ccccc4)=O, micromolar IC50=0.096957 | | Descriptor: | 4-chloro-1,3-dimethyl-N-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5Q19
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-[2-(2,4-dimethoxyphenyl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5SJX
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cn(C)nc1c4ccc(OCc3nc2ccccc2cc3)cc4)c5ccncc5, micromolar IC50=0.00070127 | | Descriptor: | 2-{[4-(1-methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)phenoxy]methyl}quinoline, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Brunner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
