1NS1
 
 | |
4BRX
 
 | | Focal Adhesion Kinase catalytic domain in complex with a diarylamino- 1,3,5-triazine inhibitor | | Descriptor: | 2-(4-(2-methoxy-4-morpholinophenylamino)-1,3,5-triazin-2-ylamino)-N-methylbenzamide, FOCAL ADHESION KINASE 1, SULFATE ION | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of Novel Diarylamino-1,3,5-Triazine Derivatives as Fak Inhibitors with Anti-Angiogenic Activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5CD1
 
 | | Structure of an asymmetric tetramer of human tRNA m1A58 methyltransferase in a complex with SAH and tRNA3Lys | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
1VP4
 
 | |
1VQZ
 
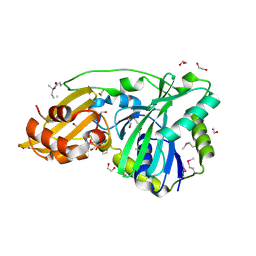 | |
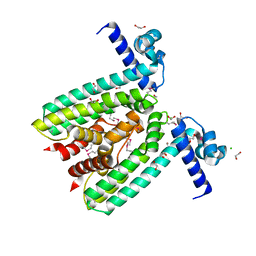
3BHQ
 
 | |
4G34
 
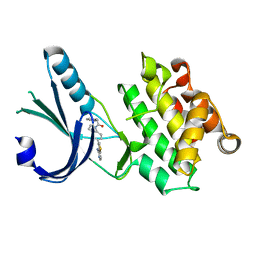 | | Crystal Structure of GSK6924 Bound to PERK (R587-R1092, delete A660-T867) at 2.70 A Resolution | | Descriptor: | 1-[5-(4-aminothieno[3,2-c]pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]-2-phenylethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
6VZI
 
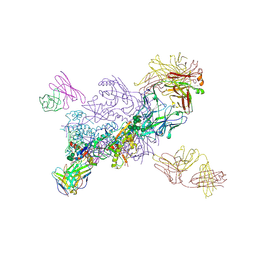 | |
4FUU
 
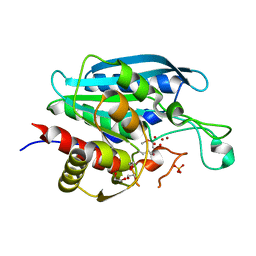 | |
4ITM
 
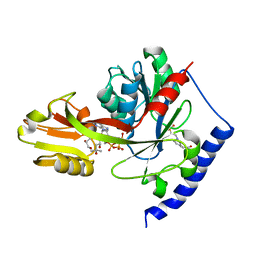 | | Crystal structure of "apo" form LpxK from Aquifex aeolicus in complex with ATP at 2.2 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1994 Å) | | Cite: | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
1XOD
 
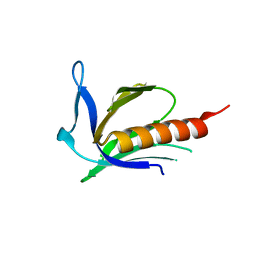 | | Crystal structure of X. tropicalis Spred1 EVH-1 domain | | Descriptor: | GLYCEROL, Spred1 | | Authors: | Harmer, N.J, Sivak, J.M, Amaya, E, Blundell, T.L. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | 1.15A Crystal structure of the X. tropicalis Spred1 EVH1 domain suggests a fourth distinct peptide-binding mechanism within the EVH1 family
Febs Lett., 579, 2005
|
|
5Y9L
 
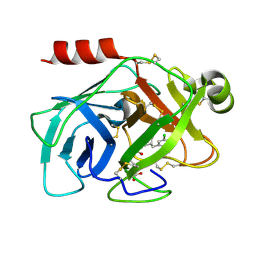 | | Human kallikrein 7 in complex with 1,3,6-trisubstituted 1,4-diazepane-7-one | | Descriptor: | 3-[2-[(3Z,6R)-6-[(5-chloranyl-2-methoxy-phenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxidanylidene-1,4-diazepan-1-yl]ethanoylamino]benzoic acid, CHLORIDE ION, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and structure-activity relationship study of 1,3,6-trisubstituted 1,4-diazepane-7-ones as novel human kallikrein 7 inhibitors
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3O14
 
 | |
3C8V
 
 | |
4NZK
 
 | |
5W4L
 
 | | Crystal structure of the non-neutralizing and ADCC-potent C11-like antibody N12-i3 in complex with HIV-1 clade A/E gp120, the CD4 mimetic M48U1, and the antibody N5-i5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody N12-i3 Fab heavy chain, Antibody N12-i3 light chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Targeting the Late Stage of HIV-1 Entry for Antibody-Dependent Cellular Cytotoxicity: Structural Basis for Env Epitopes in the C11 Region.
Structure, 25, 2017
|
|
3BDV
 
 | |
3P2C
 
 | |
1PE6
 
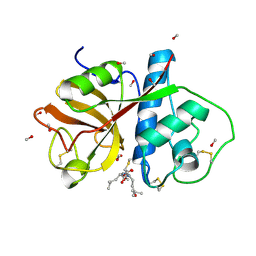 | | REFINED X-RAY STRUCTURE OF PAPAIN(DOT)E-64-C COMPLEX AT 2.1-ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE, PAPAIN | | Authors: | Yamamoto, D, Matsumoto, K, Ohishi, H, Ishida, T, Inoue, M, Kitamura, K, Mizuno, H. | | Deposit date: | 1991-05-14 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined x-ray structure of papain.E-64-c complex at 2.1-A resolution.
J.Biol.Chem., 266, 1991
|
|
5ZGP
 
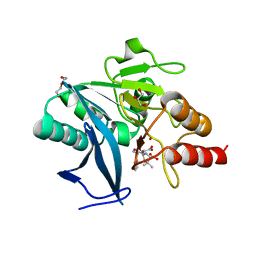 | | Crystal structure of NDM-1 at pH6.2 (Bis-Tris) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
4DOQ
 
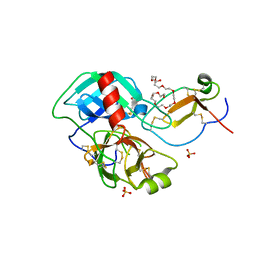 | | Crystal structure of the complex of Porcine Pancreatic Trypsin with 1/2SLPI | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Antileukoproteinase, CALCIUM ION, ... | | Authors: | Fukushima, K, Takimoto-Kamimura, M. | | Deposit date: | 2012-02-10 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure basis 1/2SLPI and porcine pancreas trypsin interaction
J.SYNCHROTRON RADIAT., 20, 2013
|
|
5C14
 
 | | Crystal structure of PECAM-1 D1D2 domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Zhou, D, Paddock, C, Newman, P, Zhu, J. | | Deposit date: | 2015-06-12 | | Release date: | 2016-01-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for PECAM-1 homophilic binding.
Blood, 127, 2016
|
|
6LIP
 
 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss0218 | | Descriptor: | (2R)-1-[3-sulfanyl-2-(sulfanylmethyl)propanoyl]pyrrolidine-2-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
5VKC
 
 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
5CCX
 
 | | Structure of the product complex of tRNA m1A58 methyltransferase with tRNA3Lys as substrate | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
