4JXS
 
 | |
8K3F
 
 | |
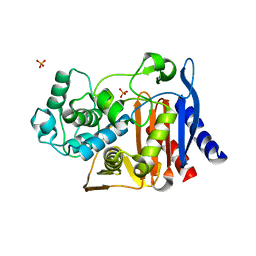
4JR5
 
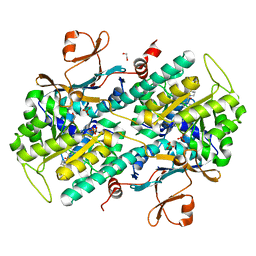 | | Structure-based Identification of Ureas as Novel Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(piperidin-1-ylsulfonyl)phenyl]-3-(pyridin-3-ylmethyl)thiourea, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Gunzner-Toste, J, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
9CT1
 
 | |
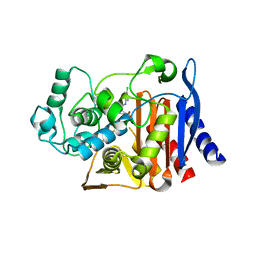
1L2S
 
 | |
4AFI
 
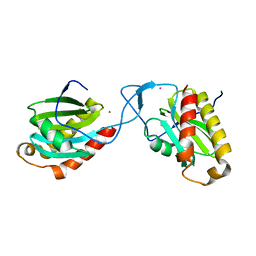 | | Complex between Vamp7 longin domain and fragment of delta-adaptin from AP3 | | Descriptor: | AP-3 COMPLEX SUBUNIT DELTA-1, VESICLE-ASSOCIATED MEMBRANE PROTEIN 7, PRASEODYMIUM ION | | Authors: | Kent, H.M, Evans, P.R, Luzio, J.P, Peden, A.A, Owen, D.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Intracellular Sorting of the Snare Vamp7 by the Ap3 Adaptor Complex
Dev.Cell, 22, 2012
|
|
8VXB
 
 | | Structure of ANT(6)-Ib from Campylobacter fetus subsp fetus complexed with hydrated streptomycin | | Descriptor: | PHOSPHATE ION, Streptomycin aminoglycoside adenylyltransferase ant(6)-Ib, [(2~{S},3~{S},4~{S},5~{R},6~{S})-2-[(2~{R},3~{R},4~{R},5~{S})-2-[(1~{R},2~{S},3~{R},4~{R},5~{S},6~{R})-2,4-bis[[azaniumylidene(azanyl)methyl]amino]-3,5,6-tris(oxidanyl)cyclohexyl]oxy-4-[bis(oxidanyl)methyl]-5-methyl-4-oxidanyl-oxolan-3-yl]oxy-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]-methyl-azanium | | Authors: | Nalam, P, Cook, P, Smith, B. | | Deposit date: | 2024-02-04 | | Release date: | 2024-09-18 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of Aminoglycoside Nucleotidyltransferase(6)-Ib From Campylobacter fetus subsp. fetus.
Proteins, 93, 2025
|
|
4KFO
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Tosteb, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4KFN
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4JNM
 
 | | Discovery of Potent and Efficacious Urea-containing Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors with Reduced CYP2C9 Inhibition Properties | | Descriptor: | 1,2-ETHANEDIOL, 1-[(6-aminopyridin-3-yl)methyl]-3-[4-(phenylsulfonyl)phenyl]urea, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Gunzner-Toste, J, Zhao, G, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Fu, B, Han, B, Ho, Y, Kley, N, Liederer, B, Lin, J, Mukadam, S, O'Brien, T, Reynolds, D.J, Sharma, G, Skelton, N, Smith, C.C, Oh, A, Wang, W, Wang, Z, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X, Bair, K.W, Dragovich, P.S. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of potent and efficacious urea-containing nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with reduced CYP2C9 inhibition properties.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7C51
 
 | |
7C50
 
 | |
4MO2
 
 | | Crystal Structure of UDP-N-acetylgalactopyranose mutase from Campylobacter jejuni | | Descriptor: | AMMONIUM ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Protsko, C, Poulin, M.B, Lowary, T.L, Sanders, D.A.R. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of a UDP-GalNAc Pyranose-Furanose Mutase: A Potential Therapeutic Target for Campylobacter jejuni Infections.
Chembiochem, 15, 2014
|
|
6C7S
 
 | | Structure of Rifampicin Monooxygenase with Product Bound | | Descriptor: | (1E,3S,4R,5S,6R,7R,8R,9S,10S,11E,13E)-15-amino-1-{[(2S)-5,7-dihydroxy-2,4-dimethyl-8-{(E)-[(4-methylpiperazin-1-yl)imino]methyl}-1,6,9-trioxo-1,2,6,9-tetrahydronaphtho[2,1-b]furan-2-yl]oxy}-7,9-dihydroxy-3-methoxy-4,6,8,10,14-pentamethyl-15-oxopentadeca-1,11,13-trien-5-yl acetate, 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Liu, L.-K, Tanner, J.J. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Evidence for Rifampicin Monooxygenase Inactivating Rifampicin by Cleaving Its Ansa-Bridge.
Biochemistry, 57, 2018
|
|
9CSZ
 
 | |
3G8E
 
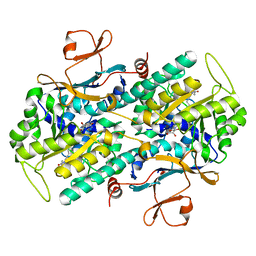 | | Crystal Structure of Rattus norvegicus Visfatin/PBEF/Nampt in Complex with an FK866-based inhibitor | | Descriptor: | 3-[(1E)-3-oxo-3-({4-[1-(phenylcarbonyl)piperidin-4-yl]butyl}amino)prop-1-en-1-yl]-1-beta-D-ribofuranosylpyridinium, Nicotinamide phosphoribosyltransferase | | Authors: | Kang, G.B, Bae, M.H, Kim, M.K, Im, I, Kim, Y.C, Eom, S.H. | | Deposit date: | 2009-02-12 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Rattus norvegicus Visfatin/PBEF/Nampt in complex with an FK866-based inhibitor
Mol.Cells, 27, 2009
|
|
4MT4
 
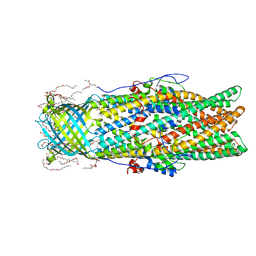 | |
4ATM
 
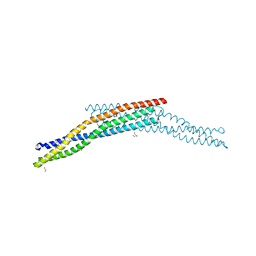 | | Crystal structure of the BAR domain of human Amphiphysin, isoform 1 at 1.8 Angstrom resolution featuring increased order at the N- terminus. | | Descriptor: | 1,2-ETHANEDIOL, AMPHIPHYSIN, GLYCEROL | | Authors: | Allerston, C.K, Krojer, T, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Crystal Structure of the Bar Domain of Human Amphiphysin, Isoform 1
To be Published
|
|
1PPR
 
 | | PERIDININ-CHLOROPHYLL-PROTEIN OF AMPHIDINIUM CARTERAE | | Descriptor: | CHLOROPHYLL A, DIGALACTOSYL DIACYL GLYCEROL (DGDG), PERIDININ, ... | | Authors: | Hofmann, E, Welte, W, Diederichs, K. | | Deposit date: | 1996-03-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae.
Science, 272, 1996
|
|
2P2V
 
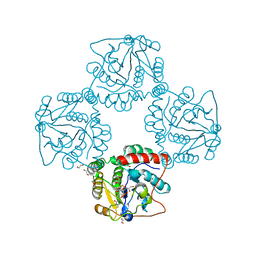 | | Crystal structure analysis of monofunctional alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase, CHLORIDE ION, ... | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
6DK4
 
 | | Crystal structure of Campylobacter jejuni peroxide stress regulator | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Ferric uptake regulation protein, MANGANESE (II) ION, ... | | Authors: | Sarvan, S, Brunzelle, J.S, Couture, J.F. | | Deposit date: | 2018-05-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of Campylobacter jejuni peroxide regulator.
FEBS Lett., 592, 2018
|
|
4BJ4
 
 | | Structure of Pseudomonas aeruginosa amidase Ampdh2 | | Descriptor: | AMPDH2, CITRATE ANION | | Authors: | Martinez-Caballero, C.S, Carrasco-Lopez, C, Artola-Recolons, C, Hermoso, J.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4BOL
 
 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with pentapeptide | | Descriptor: | AMPDH2, D-alanyl-N-[(2S,6R)-6-amino-6-carboxy-1-{[(1R)-1-carboxyethyl]amino}-1-oxohexan-2-yl]-D-glutamine, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E.E, Lastochkin, E, Zhang, W, Hellman, L.M, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4BXJ
 
 | |
3HBN
 
 | | Crystal structure PseG-UDP complex from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, GLYCEROL, UDP-sugar hydrolase, ... | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
