5HUD
 
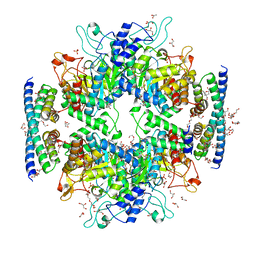 | | Non-covalent complex of and DAHP synthase and chorismate mutase from Corynebacterium glutamicum with bound transition state analog | | Descriptor: | 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
7JMH
 
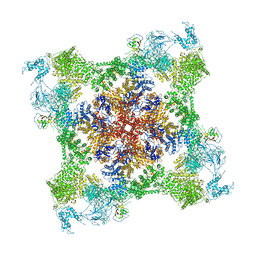 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 35 - State 4 (S4) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
6A6J
 
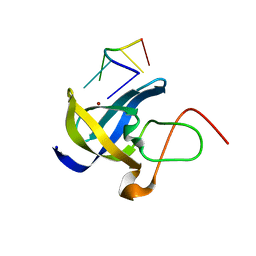 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
9HNO
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus in its iron sulfur cluster oxidized state determined by serial femtosecond crystallography | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | Deposit date: | 2024-12-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
5N8C
 
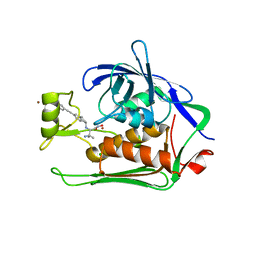 | | Crystal structure of Pseudomonas aeruginosa LpxC complexed with inhibitor | | Descriptor: | (2~{S})-3-azanyl-2-[[(1~{R})-5-[2-[4-[[2-(hydroxymethyl)imidazol-1-yl]methyl]phenyl]ethynyl]-2,3-dihydro-1~{H}-inden-1-yl]amino]-3-methyl-~{N}-oxidanyl-butanamide, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Cross, J.B, Ryan, M.D, Zhang, J, Cheng, R.K, Wood, M, Andersen, O.A, Brooks, M, Kwong, J, Barker, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based discovery of LpxC inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8BEP
 
 | |
9HNM
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus in its dark adapted and oxidized state determined by serial femtosecond crystallography | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | Deposit date: | 2024-12-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
8BED
 
 | |
3L7I
 
 | |
6TW4
 
 | | HumRadA22F in complex with compound 6 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
8BYH
 
 | |
8BEE
 
 | |
7LKJ
 
 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) | | Descriptor: | 1,2-ETHANEDIOL, Aminofutalosine deaminase, FE (III) ION | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
6DOA
 
 | |
5JHB
 
 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
9BDH
 
 | |
7JM3
 
 | | Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer | | Descriptor: | Matrix protein 1 | | Authors: | Su, Z, Pintilie, G, Selzer, L, Chiu, W, Kirkegaard, K. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer.
Plos Biol., 18, 2020
|
|
9C8T
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with AMPPNP and compound 2 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, GLYCEROL, ... | | Authors: | Wang, L, Sietsema, D. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural insight into the cGAS active site explains differences between therapeutically relevant species.
Commun Chem, 8, 2025
|
|
3LD9
 
 | |
7MOM
 
 | |
8H9X
 
 | |
3LE9
 
 | |
7MOH
 
 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 5-diphosphoinositol 1,3,4,6-tetrakisphosphate (5PP-InsP4) and phosphate in conformation B (Pi(B)) | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, PHOSPHATE ION, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
4RUX
 
 | |
8H9O
 
 | |
