6TOP
 
 | | Structure of the PorE C-terminal domain, a protein of T9SS from Porphyromonas gingivalis | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, OmpA family protein, SODIUM ION | | Authors: | Trinh, T.N, Cambillau, C, Roussel, A, Leone, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Type IX secretion system PorE C-terminal domain from Porphyromonas gingivalis in complex with a peptidoglycan fragment.
Sci Rep, 10, 2020
|
|
7KL1
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
1O49
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU85493. | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, {4-[2-ACETYLAMINO-2-(1-BIPHENYL-4-YLMETHYL-2-OXO-AZEPAN-3-YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENOXY}-ACETIC ACID | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4G
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH DPI59. | | Descriptor: | HYDROXY(1-NAPHTHYL)METHYLPHOSPHONIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
5L8C
 
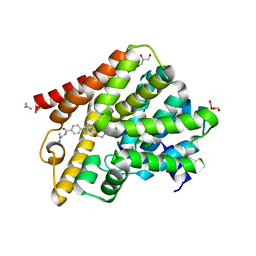 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-039 | | Descriptor: | 4-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(2-azanyl-2-oxidanylidene-ethyl)benzamide, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
1AQB
 
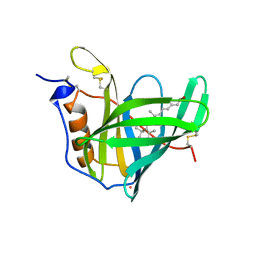 | | RETINOL-BINDING PROTEIN (RBP) FROM PIG PLASMA | | Descriptor: | CADMIUM ION, RETINOL, RETINOL-BINDING PROTEIN | | Authors: | Zanotti, G, Panzalorto, M, Marcato, A, Malpeli, G, Folli, C, Berni, R. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pig plasma retinol-binding protein at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4F1W
 
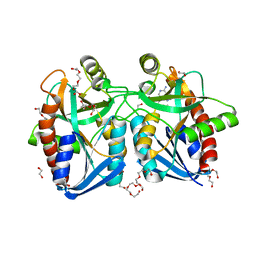 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with Adenine | | Descriptor: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with Adenine
Structure, 21, 2013
|
|
1O4A
 
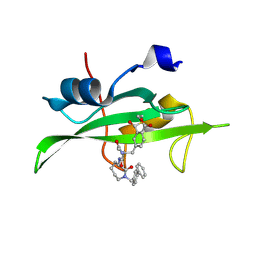 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82197. | | Descriptor: | 4-[2-ACETYLAMINO-2-(1-BIPHENYL-4-YLMETHYL-2-OXO-AZEPAN-3-YLCARBAMOYL)-ETHYL]-2-FORMYL-BENZOIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
5U35
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5AJT
 
 | | Crystal structure of ligand-free phosphoribohydrolase lonely guy from Claviceps purpurea | | Descriptor: | 1,2-ETHANEDIOL, D(-)-TARTARIC ACID, PHOSPHORIBOHYDROLASE LONELY GUY | | Authors: | Dzurova, L, Savino, S, Forneris, F. | | Deposit date: | 2015-02-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The three-dimensional structure of "Lonely Guy" from Claviceps purpurea provides insights into the phosphoribohydrolase function of Rossmann fold-containing lysine decarboxylase-like proteins.
Proteins, 83, 2015
|
|
7KP9
 
 | | asymmetric hTNF-alpha | | Descriptor: | 1-{[2-(difluoromethoxy)phenyl]methyl}-2-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-benzimidazole, Tumor necrosis factor | | Authors: | Arakaki, T.L, Abendroth, J, Fairman, J.W, Foley, A, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into the disruption of TNF-TNFR1 signalling by small molecules stabilising a distorted TNF.
Nat Commun, 12, 2021
|
|
6FV1
 
 | | Structure of human coronavirus NL63 main protease in complex with the alpha-ketoamide (S)-N-((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-2-cinnamamido-4-methylpentanamide (cinnamoyl-leucine-GlnLactam-CO-CO-NH-benzyl) | | Descriptor: | (2~{S})-4-methyl-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication Structure-based design, synthesis, and activity assessment.
J.Med.Chem., 2020
|
|
7KJ9
 
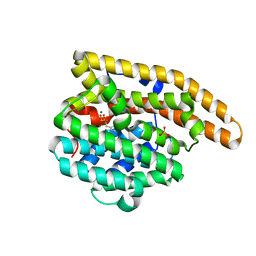 | | Wild-type epi-isozizaene synthase: complex with 3 Mg2+ and risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ronnebaum, T.A, Gardner, S.M, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
6G0D
 
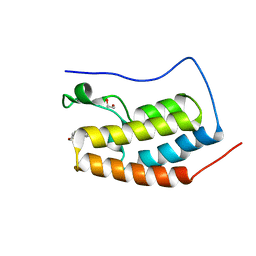 | |
7ZZB
 
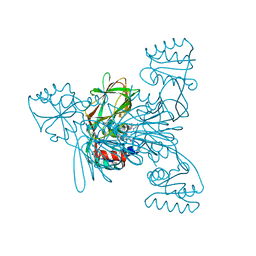 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | 2-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-[(2-azanylethylcarbamoylamino)methyl]-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]ethanoic acid, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
6C8U
 
 | | Solution structure of Musashi2 RRM1 | | Descriptor: | RNA-binding protein Musashi homolog 2 | | Authors: | Xing, M, Lan, L, Douglas, J.T, Gao, P, Hanzlik, R.P, Xu, L. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 2019
|
|
5AK8
 
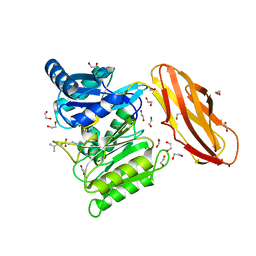 | | Structure of C351A mutant of Porphyromonas gingivalis peptidylarginine deiminase | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | Authors: | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | Deposit date: | 2015-03-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
7L4A
 
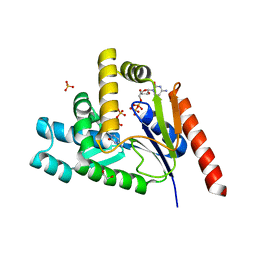 | |
7ZZ7
 
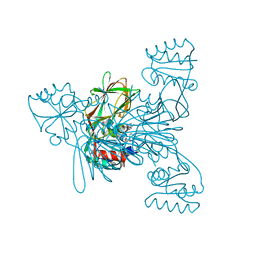 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | 2-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-[(2-azanylethanoylamino)methyl]-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]ethanoic acid, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7WLT
 
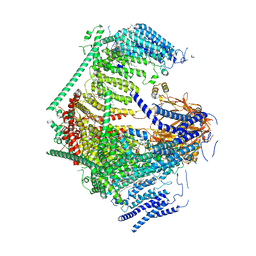 | | the Curved Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
7WT0
 
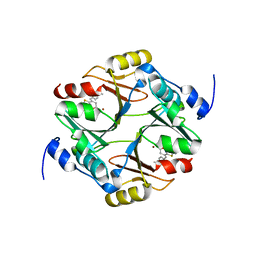 | | human glyoxalase I (with C-ter His tag) in complex with TLSC702 | | Descriptor: | (~{E})-3-(1,3-benzothiazol-2-yl)-4-(4-methoxyphenyl)but-3-enoic acid, Lactoylglutathione lyase, ZINC ION | | Authors: | Usami, M, Yokoyama, H. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human glyoxalase I and its complex with TLSC702 reveal inhibitor binding mode and substrate preference.
Febs Lett., 596, 2022
|
|
1MQI
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Fluoro-Willardiine at 1.35 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
7WT2
 
 | | human glyoxalase I in complex with TLSC702 | | Descriptor: | (~{E})-3-(1,3-benzothiazol-2-yl)-4-(4-methoxyphenyl)but-3-enoic acid, Lactoylglutathione lyase, ZINC ION | | Authors: | Usami, M, Yokoyama, H. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human glyoxalase I and its complex with TLSC702 reveal inhibitor binding mode and substrate preference.
Febs Lett., 596, 2022
|
|
5LBS
 
 | | structural basis of Zika and Dengue virus potent antibody cross-neutralization | | Descriptor: | 1,2-ETHANEDIOL, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8, SULFATE ION, ... | | Authors: | Vaney, M.C, Rouvinski, A, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
5E8T
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) | | Descriptor: | GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
