6Q7D
 
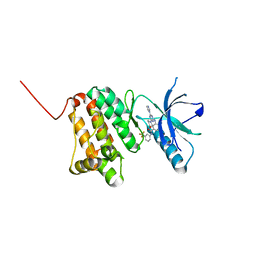 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative ATDL13 | | Descriptor: | 3-[[4-imidazol-1-yl-6-(4-oxidanylpiperidin-1-yl)-1,3,5-triazin-2-yl]amino]-4-methyl-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.978 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
8G9P
 
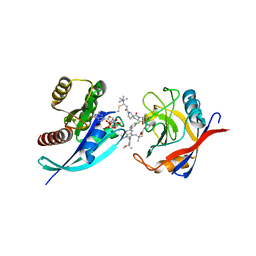 | | Tricomplex of RMC-4998, KRAS G12C, and CypA | | Descriptor: | (2S)-2-{(5S)-7-[(2E)-4-(dimethylamino)-4-methylpent-2-enoyl]-1-oxo-2,7-diazaspiro[4.4]nonan-2-yl}-N-[(1P,8S,10R,14S,21M)-22-ethyl-21-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]-3-methylbutanamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical remodeling of a cellular chaperone to target the active state of mutant KRAS.
Science, 381, 2023
|
|
6YC6
 
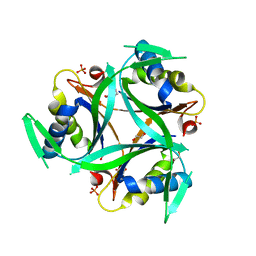 | | Structure of C. glutamicum GlnK | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, PII protein | | Authors: | Grau, F.C, Muller, Y.A. | | Deposit date: | 2020-03-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of adenylylated and unadenylylated P II protein GlnK from Corynebacterium glutamicum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8E3D
 
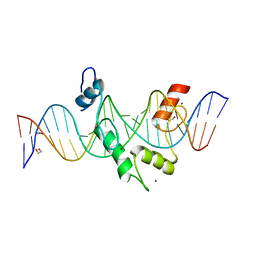 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#11) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*AP*AP*CP*C)-3'), DNA (5'-D(P*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*AP*T)-3'), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
5JZK
 
 | | The Structure of Ultra Stable Green Fluorescent Protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yong, K.J, Gunn, N.J, Scott, D.J, Griffin, M.D.W. | | Deposit date: | 2016-05-17 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Ultra-Stable, Monomeric Green Fluorescent Protein For Direct Volumetric Imaging of Whole Organs Using CLARITY.
Sci Rep, 8, 2018
|
|
6TBK
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{R},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
7PEM
 
 | | Cryo-EM structure of phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Timcenko, M, Wang, Y, Lyons, J.A, Nissen, P, Lindorff-Larsen, K. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate Transport and Specificity in a Phospholipid Flippase
To Be Published
|
|
4RE6
 
 | | Acylaminoacyl peptidase complexed with a chloromethylketone inhibitor | | Descriptor: | Acylamino-acid-releasing enzyme, CHLORIDE ION, N-[(benzyloxy)carbonyl]glycyl-N-[(2S,3R)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]glycinamide | | Authors: | Menyhard, D.K, Orgovan, Z, Szeltner, Z, Szamosi, I, Harmat, V. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Catalytically distinct states captured in a crystal lattice: the substrate-bound and scavenger states of acylaminoacyl peptidase and their implications for functionality.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9EO8
 
 | |
8E5U
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 9 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-(6-{2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}hexyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
9H16
 
 | | Crystal structure of OXA-405 apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hoff, J.F, Goudar, K.E, Hinchliffe, P, Spencer, J. | | Deposit date: | 2024-10-09 | | Release date: | 2025-09-03 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Electrostatic interactions influence diazabicyclooctane inhibitor potency against OXA-48-like beta-lactamases.
Rsc Med Chem, 2025
|
|
9AWX
 
 | | Structure of full-length amyloidogenic immunoglobulin light chain H9 in complex with 1-(1-(imidazo[1,5-a]pyrazin-8-yl)azetidin-3-yl)-3-(2-(6-methyl-4-(3-methyl-3-phenylpyrrolidin-1-yl)-2-oxopyridin-1(2H)-yl)ethyl)urea | | Descriptor: | H9 Immunoglobulin Light Chain, N-{1-[(4R)-imidazo[1,5-a]pyrazin-8-yl]azetidin-3-yl}-N'-(2-{6-methyl-4-[(3R)-3-methyl-3-phenylpyrrolidin-1-yl]-2-oxopyridin-1(2H)-yl}ethyl)urea, PHOSPHATE ION | | Authors: | Lederberg, O.L, Yan, N.L, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2024-03-05 | | Release date: | 2024-12-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Potent and Selective Pyridone-Based Small Molecule Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains.
J.Med.Chem., 67, 2024
|
|
4RF1
 
 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P63) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, S-1,2-PROPANEDIOL, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
5WGL
 
 | |
5G6V
 
 | | Crystal structure of the PCTAIRE1 kinase in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, CYCLIN-DEPENDENT KINASE 16 | | Authors: | Dixon-Clarke, S.E, Galan Bartual, S, Elkins, J, Savitsky, P, Kopec, J, Mackenzie, A, Tallant, C, Heroven, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16.
Biochem.J., 474, 2017
|
|
4QQC
 
 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Domain in Complex with FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7PG6
 
 | | Crystal Structure of PI3Kalpha in complex with the inhibitor NVP-BYL719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49943733 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
8E4Z
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 8 | | Descriptor: | (2R,3R,4R,5S)-1-(6-{[(4P)-4-(5-cyclobutyl-1,2,4-oxadiazol-3-yl)-2-nitrophenyl]amino}hexyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7MDN
 
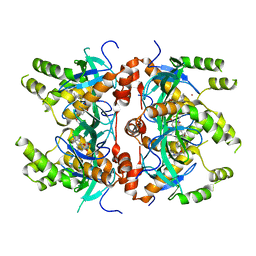 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
8E4I
 
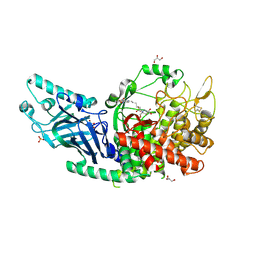 | | Co-crystal structure of Chaetomium glucosidase with compound 6 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-(6-{[(4P)-4-(5-methyl-1,2,4-oxadiazol-3-yl)-2-nitrophenyl]amino}hexyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
4QVZ
 
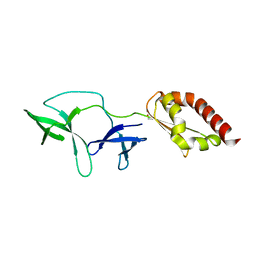 | | FMRP N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Fragile X mental retardation protein 1 | | Authors: | Myrick, L.K, Hashimoto, H, Cheng, X, Warren, S.T. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain.
Hum.Mol.Genet., 24, 2015
|
|
9H13
 
 | | Crystal structure of OXA-163 apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hoff, J.F, Goudar, K.E, Hinchliffe, P, Spencer, J. | | Deposit date: | 2024-10-09 | | Release date: | 2025-09-03 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Electrostatic interactions influence diazabicyclooctane inhibitor potency against OXA-48-like beta-lactamases.
Rsc Med Chem, 2025
|
|
7UA1
 
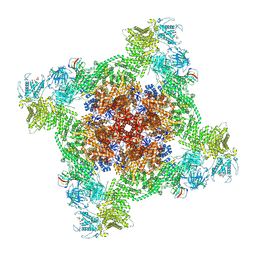 | | Structure of PKA phosphorylated human RyR2-R2474S in the closed state in the presence of ARM210 | | Descriptor: | 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment.
Sci Adv, 8, 2022
|
|
5K93
 
 | | PapD wild-type chaperone | | Descriptor: | Chaperone protein PapD | | Authors: | Sarowar, S, Li, H. | | Deposit date: | 2016-05-31 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Escherichia coli P and Type 1 Pilus Assembly Chaperones PapD and FimC Are Monomeric in Solution.
J.Bacteriol., 198, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | Descriptor: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
