1YG6
 
 | | ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Bewley, M.C, Graziano, V, Griffin, K, Flanagan, J.M. | | Deposit date: | 2005-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes.
J.Struct.Biol., 153, 2006
|
|
5G1S
 
 | | Open conformation of Francisella tularensis ClpP at 1.7 A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Diaz-Saez, L, Pankov, G, Hunter, W.N. | | Deposit date: | 2016-03-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open and compressed conformations of Francisella tularensis ClpP.
Proteins, 85, 2017
|
|
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7M1L
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, PHOSPHATE ION | | Authors: | Hall, B.M, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
5G1R
 
 | | Open conformation of Francisella tularensis ClpP at 1.9 A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Diaz-Saez, L, Hunter, W.N. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Open and compressed conformations of Francisella tularensis ClpP.
Proteins, 85, 2017
|
|
3P2L
 
 | | Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-02 | | Release date: | 2010-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis
To be Published
|
|
7MK5
 
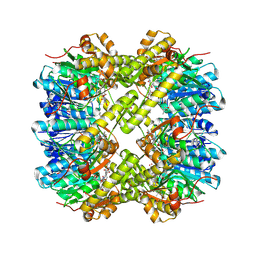 | | Crystal structure of Escherichia coli ClpP covalently inhibited by clipibicyclene | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-[(1E)-3-{[(2E,4E,6E,8S)-8-hydroxy-4-methyldeca-2,4,6-trienoyl]amino}-3-oxoprop-1-en-1-yl]azete-1(2H)-carboxylic acid, ACETATE ION, ... | | Authors: | Culp, E.J, Sychantha, D, Hobson, C, Pawlowski, A.J, Prehna, G, Wright, G.D. | | Deposit date: | 2021-04-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | ClpP inhibitors are produced by a widespread family of bacterial gene clusters.
Nat Microbiol, 7, 2022
|
|
4RYF
 
 | | ClpP1/2 heterocomplex from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, MALONATE ION, SODIUM ION | | Authors: | Dahmen, M, Vielberg, M.-T, Groll, M, Sieber, S.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of the caseinolytic protease ClpP1/2 heterocomplex from Listeria monocytogenes.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3QWD
 
 | | Crystal structure of ClpP from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | Authors: | Geiger, S.R, Boettcher, T, Sieber, S.A, Cramer, P. | | Deposit date: | 2011-02-28 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Conformational Switch Underlies ClpP Protease Function.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3Q7H
 
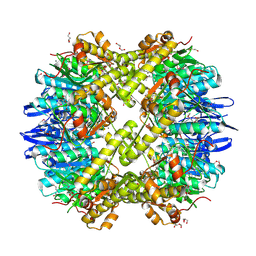 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
4U0H
 
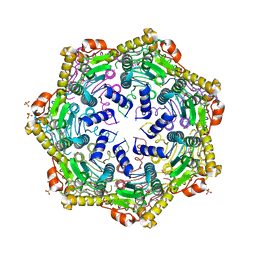 | | Crystal Structure of M. tuberculosis ClpP1P1 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, SULFATE ION | | Authors: | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2479 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U0G
 
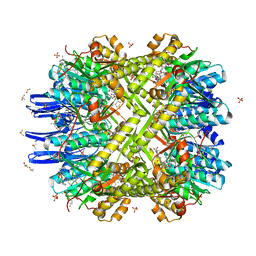 | | Crystal Structure of M. tuberculosis ClpP1P2 bound to ADEP and agonist | | Descriptor: | ADEP-2B5Me, ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, ... | | Authors: | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1978 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5G1Q
 
 | |
4GM2
 
 | | The crystal structure of a peptidase from plasmodium falciparum | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | El Bakkouri, M, Jung, P, Wernimont, A.K, Calmettes, C, Hui, R, Houry, W.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-15 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Inactive Subunit of the Apicoplast-localized Caseinolytic Protease Complex of Plasmodium falciparum.
J.Biol.Chem., 288, 2013
|
|
4HNK
 
 | | Crystal structure of an Enzyme | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL | | Authors: | El Bakkouri, M, Calmettes, C, Wernimont, A.K, Houry, W.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-19 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Inactive Subunit of the Apicoplast-localized Caseinolytic Protease Complex of Plasmodium falciparum.
J.Biol.Chem., 288, 2013
|
|
6HWM
 
 | | Structure of Thermus thermophilus ClpP in complex with bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QYF
 
 | |
6H23
 
 | | Crystal structure of the hClpP Y118A mutant with an activating small molecule | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Kick, L.M, Sieber, S.A, Schneider, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Selective Activation of Human Caseinolytic Protease P (ClpP).
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6IW7
 
 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | (2S)-2-benzamido-4-methyl-pentanoic acid, 4-[(4-fluoro-2-methyl-3H-indol-5-yl)oxy]-6-methoxy-7-[3-(pyrrolidin-1-yl)propoxy]quinazoline, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2018-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.692121 Å) | | Cite: | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease
To Be Published
|
|
4JCQ
 
 | | ClpP1 from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zeiler, E, List, A, Alte, F, Gersch, M, Wachtel, R, Groll, M, Sieber, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JCT
 
 | | ClpP2 from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zeiler, E, List, A, Alte, F, Gersch, M, Wachtel, R, Groll, M, Sieber, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6X60
 
 | | ClpP2 from Chlamydia trachomatis with resolved handle loop | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | Azadmanesh, J, Struble, L.R, Seleem, M.A, Ouellette, S, Conda-Sheridan, M, Borgstahl, G.E.O. | | Deposit date: | 2020-05-27 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | ClpP2 from Chlamydia trachomatis with resolved handle loop
To Be Published
|
|
