1OU5
 
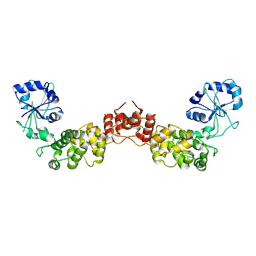 | | Crystal structure of human CCA-adding enzyme | | Descriptor: | tRNA CCA-adding enzyme | | Authors: | Augustin, M.A, Reichert, A.S, Betat, H, Huber, R, Moerl, M, Steegborn, C. | | Deposit date: | 2003-03-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the Human CCA-adding Enzyme: Insights into Template-independent Polymerization
J.Mol.Biol., 328, 2003
|
|
7Z27
 
 | |
5URN
 
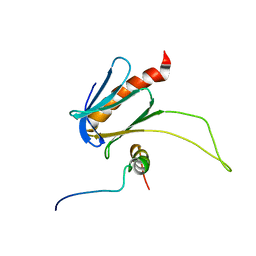 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the transactivation domain 1 of p65 | | Descriptor: | RNA polymerase II transcription factor B subunit 1, Transcription factor p65 | | Authors: | Lecoq, L, Omichinski, J.G, Raiola, L, Cyr, N, Chabot, P, Arseneault, G, Legault, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
4OWR
 
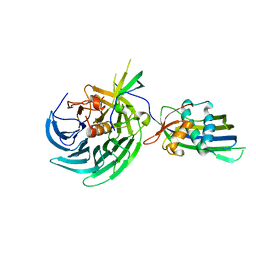 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | Descriptor: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | Authors: | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | Deposit date: | 2014-02-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6G2Q
 
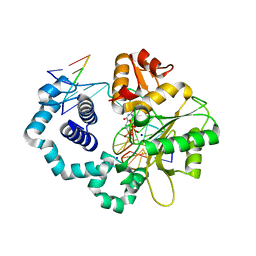 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL (S-isomer), beta, gamma dTTP analogue | | Descriptor: | CHLORIDE ION, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
2B7E
 
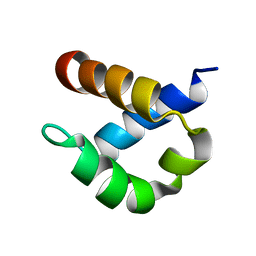 | | First FF domain of Prp40 Yeast Protein | | Descriptor: | Pre-mRNA processing protein PRP40 | | Authors: | Gasch, A, Wiesner, S, Martin-Malpartida, P, Ramirez-Espain, X, Ruiz, L, Macias, M.J. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains.
J.Biol.Chem., 281, 2006
|
|
1TCO
 
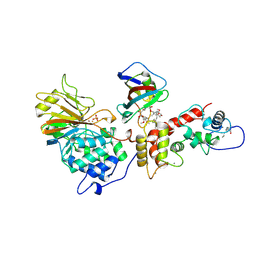 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
4RT3
 
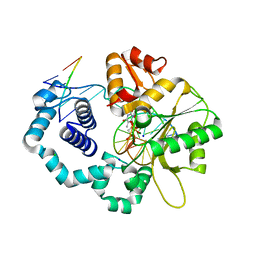 | | Ternary complex crystal structure of DNA polymerase Beta with (alpha, beta)-NH-(beta,gamma)-CH2-dTTP | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(R)-hydroxy(phosphonomethyl)phosphoryl]amino}phosphoryl]-3,4-dihydrothymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2014-11-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Two Scaffolds from Two Flips: (alpha, beta)/(beta, gamma) CH2/NH "Met-Im" Analogues of dTTP.
Org.Lett., 17, 2015
|
|
8KIA
 
 | |
7Y5M
 
 | |
4C0H
 
 | |
7YZV
 
 | |
1O6W
 
 | |
3DOM
 
 | | Crystal Structure of the complex between Tfb5 and the C-terminal domain of Tfb2 | | Descriptor: | RNA polymerase II transcription factor B subunit 2, RNA polymerase II transcription factor B subunit 5 | | Authors: | Kainov, D.E, Cavarelli, J, Egly, J.M, Poterszman, A. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for group A trichothiodystrophy
Nat.Struct.Mol.Biol., 15, 2008
|
|
4RT2
 
 | | Ternary complex crystal structure of DNA polymerase Beta with (alpha,beta)-CH2-(beta,gamma)-NH-dTTP | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]methyl}phosphoryl]-3,4-dihydrothymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2014-11-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Two Scaffolds from Two Flips: (alpha, beta)/(beta, gamma) CH2/NH "Met-Im" Analogues of dTTP.
Org.Lett., 17, 2015
|
|
3DGP
 
 | | Crystal Structure of the complex between Tfb5 and the C-terminal domain of Tfb2 | | Descriptor: | RNA polymerase II transcription factor B subunit 2, RNA polymerase II transcription factor B subunit 5 | | Authors: | Kainov, D.E, Cavarelli, J, Egly, J.M, Poterszman, A. | | Deposit date: | 2008-06-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for group A trichothiodystrophy
Nat.Struct.Mol.Biol., 15, 2008
|
|
4TQR
 
 | |
4TQS
 
 | |
5V5I
 
 | |
4I1T
 
 | |
4KHQ
 
 | | Ternary complex of RB69 mutant L415F wit DUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Clausen, A.R, Pedersen, L.C. | | Deposit date: | 2013-05-01 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Structure-function analysis of ribonucleotide bypass by B family DNA replicases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6BEL
 
 | | Ternary complex crystal structure of DNA polymerase Beta with R-isomer of beta-gamma-CHF-dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-deoxy-5'-O-[(R)-{[(R)-[(R)-fluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2017-10-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR3
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CBr2, beta, gamma dATP analogue | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-[dibromo(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]adenosine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*TP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CTM
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL(R-isomer), beta, gamma dTTP analogue | | Descriptor: | 5'-O-[(R)-{[(R)-[(R)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CTN
 
 | |
