7WM2
 
 | | Cryo-EM structure of AKT1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Dongliang, L, Zijie, Z, Yannan, Q, Yuyue, T, Huaizong, S. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis for the regulation mechanism of a hyperpolarization-activated K+ channel AKT1 by AtKC1
To Be Published
|
|
7QSC
 
 | | GTPase IN COMPLEX WITH GDP.MGF3- | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho GTPase-activating protein 1, ... | | Authors: | Baumann, P, Jin, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | GTPase IN COMPLEX WITH GDP.MGF3-
To Be Published
|
|
7TIJ
 
 | |
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THS
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2S)-butane-1,2-diol, (6S,9R,20R,23S)-N-{[4-(aminomethyl)phenyl]methyl}-20-[(benzenesulfonyl)amino]-3,13,21-trioxo-2,6,9,14,22-pentaazatetracyclo[23.2.2.2~6,9~.2~15,18~]tritriaconta-1(27),15,17,25,28,30-hexaene-23-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7WL3
 
 | | CVB5 expended empty particle | | Descriptor: | Capsid protein, Genome polyprotein | | Authors: | Yang, P, Wang, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.
J.Virol., 96, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7QRN
 
 | | Crystal structure of Ovalbumin-related protein X (OVAX) complexed with fondaparinux | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Ovalbumin-related protein X, ... | | Authors: | Coste, F, Bruneau, G, Rehault-Godbert, S. | | Deposit date: | 2022-01-11 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Antimicrobial Proteins and Peptides in Avian Eggshell: Structural Diversity and Potential Roles in Biomineralization.
Front Immunol, 13, 2022
|
|
7QRM
 
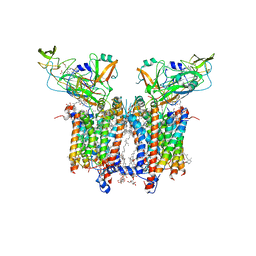 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.7 A resolution | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Sarewicz, M, Szwalec, M, Indyka, P, Rawski, M, Pintscher, S, Pietras, R, Mielecki, B, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
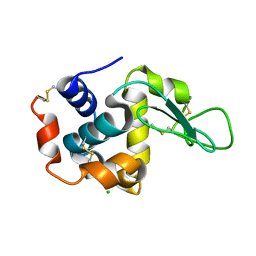
7WKR
 
 | |
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
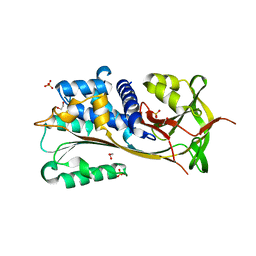
7QR2
 
 | |
7QRA
 
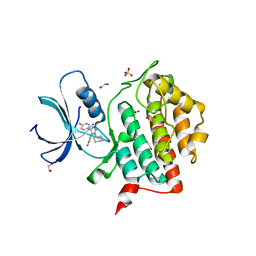 | | Crystal structure of CK1 delta in complex with VN725 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-cyclohexyl-5-(4-fluorophenyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Nemec, V, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QR9
 
 | | Crystal structure of CK1 delta in complex with PK-09-82 | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(4-fluorophenyl)-3-(pyridin-4-ylmethyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QRB
 
 | | Crystal structure of CK1 delta in complex with PK-09-129 | | Descriptor: | 3-(dimethylamino)-~{N}-[4-[4-(4-fluorophenyl)-5-(1~{H}-pyrrolo[2,3-b]pyridin-4-yl)imidazol-1-yl]cyclohexyl]propane-1-sulfonamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WKI
 
 | |
7QR3
 
 | | Chimpanzee CPEB3 HDV-like ribozyme | | Descriptor: | GLYCEROL, POTASSIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
7WKD
 
 | | TRH-TRHR G protein complex | | Descriptor: | Gq, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Y, Cai, H, You, C, He, X, Yuan, Q, Jiang, H, Cheng, X, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into ligand binding and activation of the human thyrotropin-releasing hormone receptor.
Cell Res., 32, 2022
|
|
7WKG
 
 | | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with erucic acid | | Descriptor: | (Z)-docos-13-enoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with erucic acid
To Be Published
|
|
7WKB
 
 | | The 0.81 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with docosahexaenoic acid | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | The 0.81 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with docosahexaenoic acid
To Be Published
|
|
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
7QQ2
 
 | |
