5GSQ
 
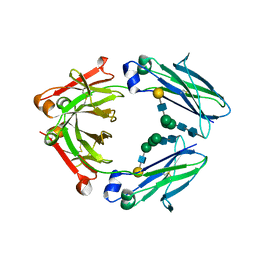 | | Crystal structure of IgG Fc with a homogeneous glycoform and Antibody-Dependent Cellular Cytotoxicity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, C.-L, Hsu, J.-C, Lin, C.-W, Wu, C.-Y, Wong, C.-H, Ma, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a Homogeneous IgG-Fc Glycoform with the N-Glycan Designed to Maximize the Antibody Dependent Cellular Cytotoxicity
ACS Chem. Biol., 12, 2017
|
|
2HRR
 
 | |
2HRQ
 
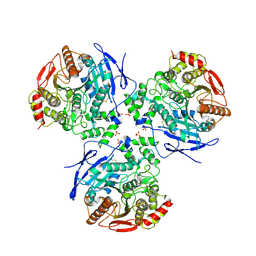 | | Crystal structure of Human Liver Carboxylesterase 1 (hCE1) in covalent complex with the nerve agent Soman (GD) | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (R)-METHYLPHOSPHINATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Liver carboxylesterase 1, ... | | Authors: | Fleming, C.D, Redinbo, M.R. | | Deposit date: | 2006-07-20 | | Release date: | 2007-05-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Human Carboxylesterase 1 in Covalent Complexes with the Chemical Warfare Agents Soman and Tabun.
Biochemistry, 46, 2007
|
|
2P59
 
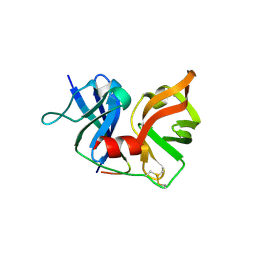 | | Crystal Structure of Hepatitis C Virus NS3.4A protease | | Descriptor: | (2S,3AS,7AS)-1-[(2S)-2-{[(2S)-2-CYCLOHEXYL-2-({[(2R)-4-NITRO-2H-PYRROL-2-YL]CARBONYL}AMINO)ACETYL]AMINO}-3,3-DIMETHYLBUTANOYL]-N-{(1S)-1-[(1R)-2-(CYCLOPROPYLAMINO)-1-HYDROXY-2-OXOETHYL]BUTYL}OCTAHYDRO-1H-INDOLE-2-CARBOXAMIDE, NS3, peptide | | Authors: | Perni, R.B, Wei, Y. | | Deposit date: | 2007-03-14 | | Release date: | 2008-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibitors of hepatitis C virus NS3.4A protease. Effect of P4 capping groups on inhibitory potency and pharmacokinetics.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3CWX
 
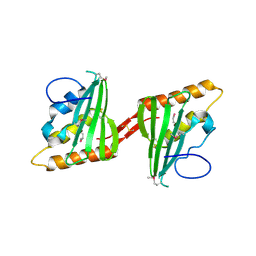 | | Crystal structure of cagd from helicobacter pylori pathogenicity island | | Descriptor: | protein CagD | | Authors: | Cendron, L, Zanotti, G, Angelini, A, Barison, N, Couturier, M, Stein, M. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion.
J.Mol.Biol., 386, 2009
|
|
9B13
 
 | |
4TX3
 
 | | Complex of the X-domain and OxyB from Teicoplanin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, OxyB protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Peschke, M, Haslinger, K, Cryle, M.J. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis.
Nature, 521, 2015
|
|
4I5C
 
 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-3-[(3R)-3-(pyrrolo[2,3-b][1,2,3]triazolo[4,5-d]pyridin-1(6H)-yl)piperidin-1-yl]propanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazolo-pyrrolopyridines as inhibitors of Janus kinase 1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4E5W
 
 | | JAK1 kinase (JH1 domain) in complex with compound 26 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine-protein kinase JAK1, [4-(imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl)piperidin-1-yl][(2S)-1-(propan-2-yl)pyrrolidin-2-yl]methanone | | Authors: | Murray, J.M. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4F08
 
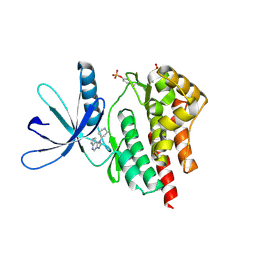 | |
4F09
 
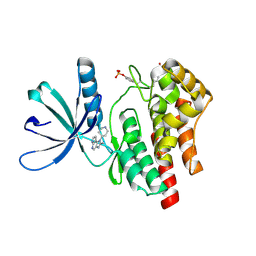 | |
4DXD
 
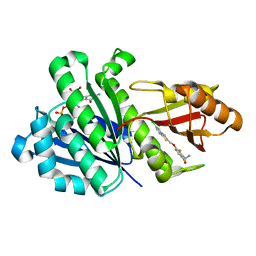 | | Staphylococcal Aureus FtsZ in complex with 723 | | Descriptor: | 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lu, J, Soisson, S.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Restoring methicillin-resistant Staphylococcus aureus susceptibility to beta-lactam antibiotics.
Sci Transl Med, 4, 2012
|
|
4E6Q
 
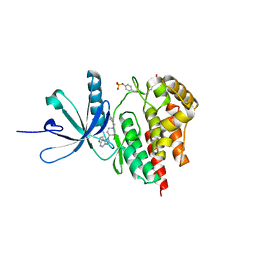 | | JAK2 kinase (JH1 domain) triple mutant in complex with compound 12 | | Descriptor: | 1-(1-benzylpiperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK2 | | Authors: | Murray, J.M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4EHZ
 
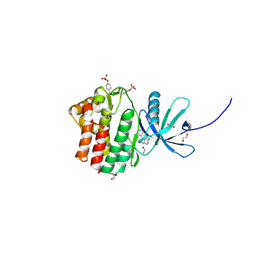 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-1-(piperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4E6D
 
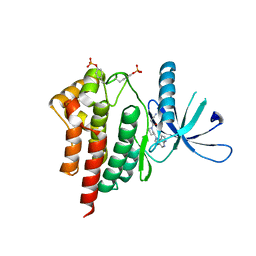 | | JAK2 kinase (JH1 domain) triple mutant in complex with compound 7 | | Descriptor: | 3-[(3R)-3-(imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl)piperidin-1-yl]-3-oxopropanenitrile, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Murray, J.M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
5PGU
 
 | |
5PGX
 
 | |
5PGZ
 
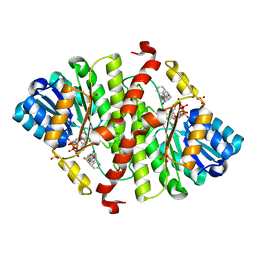 | | CRYSTAL STRUCTURE OF MURINE 11BETA- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
3NMU
 
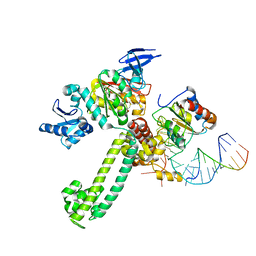 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-06-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3NVK
 
 | | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Xue, S, Wang, R, Li, H. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3NVM
 
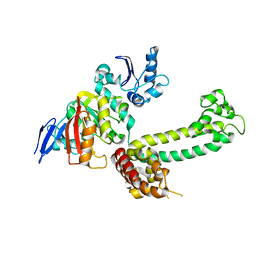 | |
5NPP
 
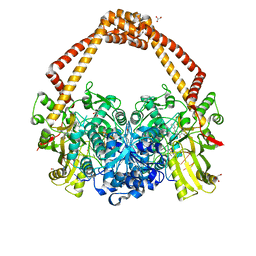 | | 2.22A STRUCTURE OF THIOPHENE2 AND GSK945237 WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DIMETHYL SULFOXIDE, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Chan, P.F, Stavenger, R.A. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NPK
 
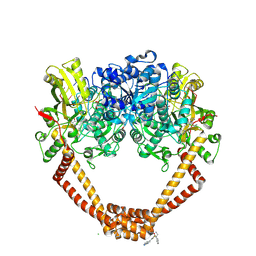 | | 1.98A STRUCTURE OF THIOPHENE1 WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit B,DNA gyrase subunit B,DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Chan, P.F, Stavenger, R.A. | | Deposit date: | 2017-04-17 | | Release date: | 2017-07-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5PGY
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
3NVI
 
 | | Structure of N-terminal truncated Nop56/58 bound with L7Ae and box C/D RNA | | Descriptor: | 50S ribosomal protein L7Ae, NOP5/NOP56 related protein, RNA (5'-R(*CP*UP*CP*UP*GP*AP*CP*CP*GP*AP*AP*AP*GP*GP*CP*GP*UP*GP*AP*UP*GP*AP*GP*C)-3') | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
