5M9J
 
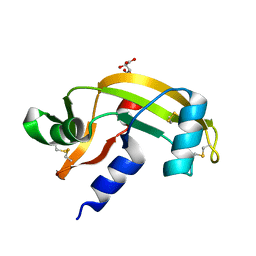 | | Human angiogenin PD variant K60E | | Descriptor: | Angiogenin, L(+)-TARTARIC ACID | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5MTK
 
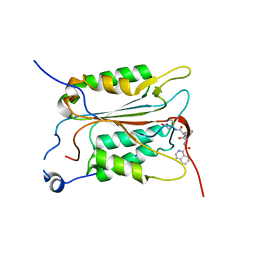 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
5MK8
 
 | |
5M9S
 
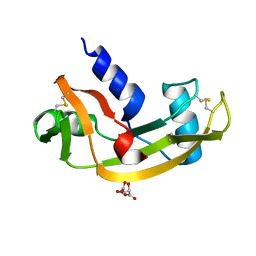 | | Human angiogenin ALS variant V103I | | Descriptor: | Angiogenin, D(-)-TARTARIC ACID | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
7KS4
 
 | |
5MK7
 
 | |
7KJJ
 
 | | Reconstructed ancestor of HIUases and Transthyretins | | Descriptor: | 1,2-ETHANEDIOL, 3,5,3',5'-TETRAIODO-L-THYRONINE, TTR ancestor | | Authors: | Nagem, R.A.P, Bleicher, L, Costa, M.A.F. | | Deposit date: | 2020-10-26 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Reenacting the Birth of a Function: Functional Divergence of HIUases and Transthyretins as Inferred by Evolutionary and Biophysical Studies.
J.Mol.Evol., 89, 2021
|
|
7NAY
 
 | | Crystal structure of lactate dehydrogenase from Selenomonas ruminantium with NADH. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, L-lactate dehydrogenase, ... | | Authors: | Bertrand, Q, Robin, A, Girard, E, Madern, D. | | Deposit date: | 2021-01-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
7MGJ
 
 | |
7MGK
 
 | |
7M0J
 
 | |
7LJR
 
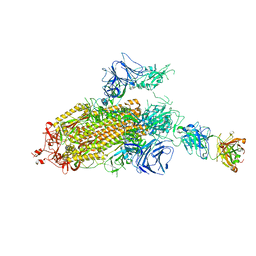 | | SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab | | Descriptor: | Fab DH1043 heavy chain, Fab DH1043 light chain, Spike glycoprotein | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates.
Biorxiv, 2021
|
|
7LWS
 
 | |
4QFV
 
 | |
2B54
 
 | | Human cyclin dependent kinase 2 (CKD2)complexed with DIN-232305 | | Descriptor: | 6-(3,4-DIHYDROXYBENZYL)-3-ETHYL-1-(2,4,6-TRICHLOROPHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4(5H)-ONE, Cell division protein kinase 2 | | Authors: | Chang, C.-C. | | Deposit date: | 2005-09-27 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological evaluation of 1-aryl-4,5-dihydro-1h-pyraxolo[3,4-d]pyrimidin-4-one inhibitors of cyclin dependent kinases
J.Med.Chem., 47, 2004
|
|
1YFF
 
 | | STRUCTURE OF HUMAN CARBONMONOXYHEMOGLOBIN C (BETA E6K): TWO QUATERNARY STATES (R2 and R3) IN ONE CRYSTAL | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L, Patskovsky, Y, Hirsch, R.E, Almo, S.C. | | Deposit date: | 2004-12-31 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Quaternary Transitions in Human Liganded Hemoglobin
To be Published
|
|
3BJL
 
 | |
1G9F
 
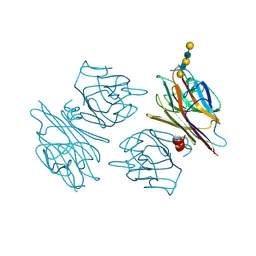 | | CRYSTAL STRUCTURE OF THE SOYBEAN AGGLUTININ IN A COMPLEX WITH A BIANTENNARY BLOOD GROUP ANTIGEN ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Buts, L, Hamelryck, T.W, Dao-Thi, M.-H, Loris, R, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-23 | | Release date: | 2001-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
5C1G
 
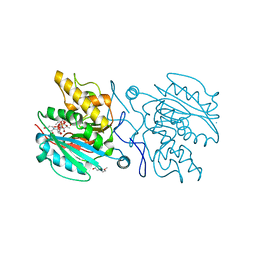 | | Crystal structure of ABBA + UDP + DI | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GALACTOSE-URIDINE-5'-DIPHOSPHATE, Histo-blood group ABO system transferase, ... | | Authors: | Gagnon, S. | | Deposit date: | 2015-06-13 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | High Resolution Structures of the Human ABO(H) Blood Group Enzymes in Complex with Donor Analogs Reveal That the Enzymes Utilize Multiple Donor Conformations to Bind Substrates in a Stepwise Manner.
J.Biol.Chem., 290, 2015
|
|
5BXC
 
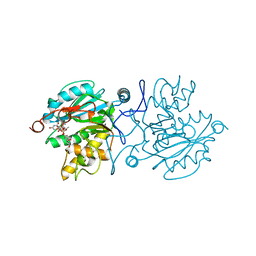 | | Crystal structure of GTA + UDP + DI | | Descriptor: | Histo-blood group ABO system transferase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Gagnon, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Structures of the Human ABO(H) Blood Group Enzymes in Complex with Donor Analogs Reveal That the Enzymes Utilize Multiple Donor Conformations to Bind Substrates in a Stepwise Manner.
J.Biol.Chem., 290, 2015
|
|
8I0C
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S0703 | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, He, S, Liu, Y, Fang, P, Sun, H. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
5CJ3
 
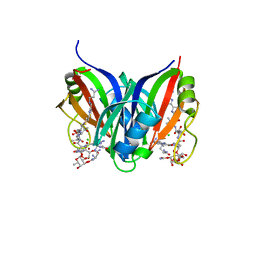 | | Crystal structure of the zorbamycin binding protein (ZbmA) from Streptomyces flavoviridis with zorbamycin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Zbm binding protein, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Rudolf, J.D, Ma, M, Chang, C.-Y, Lohman, J.R, Yang, D, Shen, B, Enzyme Discovery for Natural Product Biosynthesis, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-13 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6499 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
5CNX
 
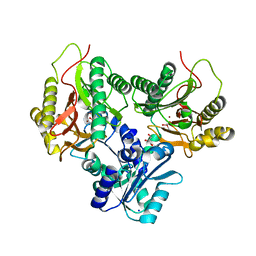 | | Crystal structure of Xaa-Pro aminopeptidase from Escherichia coli K12 | | Descriptor: | Aminopeptidase YpdF, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-18 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases
Proteins, 2018
|
|
5D2Y
 
 | |
5D7Y
 
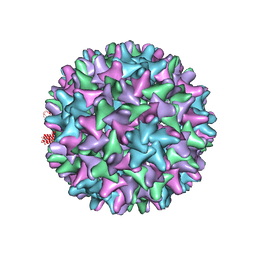 | | Crystal structure of Hepatitis B virus T=4 capsid in complex with the allosteric modulator HAP18 | | Descriptor: | Capsid protein, methyl (4R)-4-(2-chloro-4-fluorophenyl)-6-{[4-(3-hydroxypenta-1,4-diyn-3-yl)piperidin-1-yl]methyl}-2-(pyridin-2-yl)-1,4-dihydropyrimidine-5-carboxylate | | Authors: | Venkatakrishnan, B, Katen, S.P, Zlotnick, A. | | Deposit date: | 2015-08-14 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.894 Å) | | Cite: | Hepatitis B Virus Capsids Have Diverse Structural Responses to Small-Molecule Ligands Bound to the Heteroaryldihydropyrimidine Pocket.
J.Virol., 90, 2016
|
|
