7T0B
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2g | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-5-butyl-4-({1-[(4-fluorophenyl)methyl]-1H-imidazol-5-yl}methyl)-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
8QFZ
 
 | | TSLP-Bicycle complex | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, CYS-HIS-TRP-LEU-GLU-ASN-CYS-TRP-ARG-GLY-PHE-CYS, Thymic stromal lymphopoietin | | Authors: | Petersen, J. | | Deposit date: | 2023-09-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Characterization of a Bicyclic Peptide (Bicycle) Binder to Thymic Stromal Lymphopoietin.
J.Med.Chem., 67, 2024
|
|
7T0E
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2b | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-butyl-1-(3-chlorophenyl)piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
6GJ6
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH 18 | | Descriptor: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A, Phan, J. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
9HBL
 
 | | SmsC2B2 complex from M. jannaschii (monoclinic form) | | Descriptor: | 1,2-ETHANEDIOL, Iron-sulfur cluster assembly SufBD family protein MJ0034, SULFATE ION, ... | | Authors: | Mechaly, A.E, Dussouchaud, M, Haouz, A, Betton, J.M, Barras, F. | | Deposit date: | 2024-11-07 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Ancestral [Fe-S] biogenesis system SMS has a unique mechanism of cluster assembly and sulfur utilization.
Plos Biol., 23, 2025
|
|
7XXN
 
 | | HapR Quadruple mutant, bound to Qstatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, GLYCEROL, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
6EG3
 
 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
7T09
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2d | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 1,2-ETHANEDIOL, 4-{[5-({(2S)-2-butyl-5-oxo-4-[3-(trifluoromethoxy)phenyl]piperazin-1-yl}methyl)-1H-imidazol-1-yl]methyl}benzonitrile, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
8VTY
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with ciprofloxacin and protein Y at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 16S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
7T0A
 
 | | Cryptococcus neoformans protein farnesyltransferase co-crystallized with FPP and inhibitor 2f | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-butyl-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7BAY
 
 | |
6O7S
 
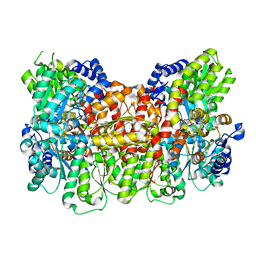 | |
6RDP
 
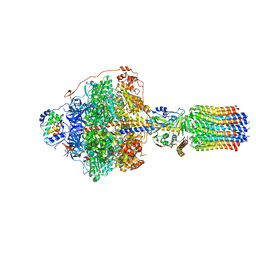 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1C, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
8C23
 
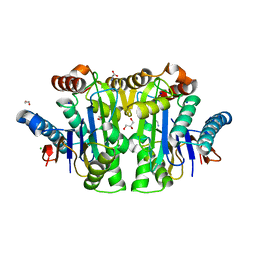 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (monoclinic form M200T#m) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
7T08
 
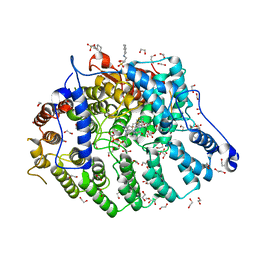 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2q | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 1,2-ETHANEDIOL, 4-{[5-({(2S)-2-butyl-5-oxo-4-[3-(trifluoromethoxy)phenyl]piperazin-1-yl}methyl)-1H-imidazol-1-yl]methyl}-2-fluorobenzonitrile, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7XGT
 
 | |
4U8I
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
6RDG
 
 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 3, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDS
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1D, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
7BF2
 
 | | Ca2+-Calmodulin in complex with human muscle form creatine kinase peptide in extended 1:2 binding mode | | Descriptor: | CALCIUM ION, Calmodulin-1, Creatine kinase M-type | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
9BXN
 
 | | OvoM from Sulfuricurvum sp. isolate STB_99, an ovothiol-biosynthetic N-methyltransferase in complex with 5-thiohistidine and SAH | | Descriptor: | 1,2-ETHANEDIOL, 5-sulfanyl-L-histidine, 5-thiohistidine N-methyltransferase OvoM, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-05-22 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of SAM-dependent N-methyltransferases involved in ovoselenol and ovothiol biosynthesis.
Structure, 33, 2025
|
|
6Y50
 
 | | 5'domain of human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
8H63
 
 | | Crystal structure of Internalin A from Listeria monocytogenes with nanobody VHH10 bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Nagatoish, S, Tsumoto, K. | | Deposit date: | 2022-10-15 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Anti-InlA single-domain antibodies that inhibit the cell invasion of Listeria monocytogenes.
J.Biol.Chem., 299, 2023
|
|
9BXM
 
 | |
8DHN
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-2-(5-methyl-1,2,4-oxadiazol-3-yl)pyridin-4(1H)-one | | Descriptor: | (2P)-6-bromo-3-hydroxy-2-(5-methyl-1,2,4-oxadiazol-3-yl)pyridin-4(1H)-one, MANGANESE (II) ION, PA endonuclease | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-27 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
