6UEG
 
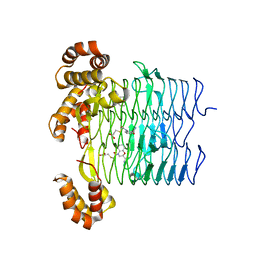 | | Pseudomonas aeruginosa LpxA Complex Structure with Ligand | | Descriptor: | 3-({2-[(2R)-2-carbamoyl-2,3-dihydro-4H-1,4-benzoxazin-4-yl]-2-oxoethyl}sulfanyl)propanoic acid, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CALCIUM ION | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UFA
 
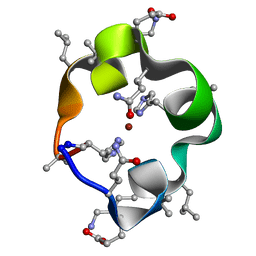 | | S4 symmetric peptide design number 1, Tim zinc-bound form | | Descriptor: | S4-1, Tim, Zinc-bound form, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6HOP
 
 | |
8Q10
 
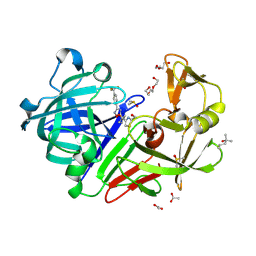 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-(hydroxymethyl)thiazol-4-yl)-5-(3-(4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-125) | | Descriptor: | (3~{R},5~{R})-3-[2-(hydroxymethyl)-1,3-thiazol-4-yl]-5-[3-[4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
7PUP
 
 | | INOSITOL 1,3,4-TRISPHOSPHATE 5/6-KINASE 4 from Arabidopsis thaliana (AtITPK4) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol 1,3,4-trisphosphate 5/6-kinase 4 | | Authors: | Whitfield, H, He, S, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Diversification in the inositol tris/tetrakisphosphate kinase (ITPK) family: crystal structure and enzymology of the outlier AtITPK4.
Biochem.J., 480, 2023
|
|
8DDF
 
 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
8Q0X
 
 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-((methyl(propyl)amino)methyl)thiazol-4-yl)-5-(3-(4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-087) | | Descriptor: | (3~{R},5~{R})-3-[2-[[methyl(propyl)amino]methyl]-1,3-thiazol-4-yl]-5-[3-[4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
6HNB
 
 | | Crystal structure of aminotransferase Aro8 from Candida albicans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aromatic amino acid aminotransferase I, CHLORIDE ION, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
9IK4
 
 | |
9MIF
 
 | | Crystal structure of the VRC01-class antibody 9C09, derived from GT1.1 vaccination, in complex with eOD-GT8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9C09 Fab heavy chain, 9C09 Fab light chain, ... | | Authors: | Agrawal, S, Wilson, I.A. | | Deposit date: | 2024-12-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Precise targeting of HIV broadly neutralizing antibody precursors in humans.
Science, 389, 2025
|
|
6P7J
 
 | |
7VLU
 
 | | Structure of SUR2A in complex with Mg-ATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
8UN4
 
 | |
7VLS
 
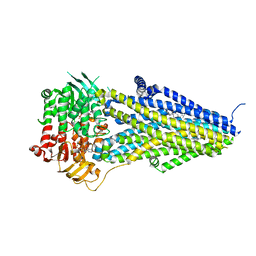 | | Structure of SUR2B in complex with MgATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
6Y15
 
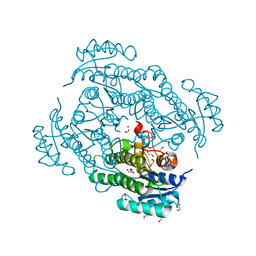 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant T102E_Q126K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
5K8G
 
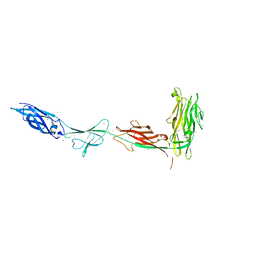 | |
6DK2
 
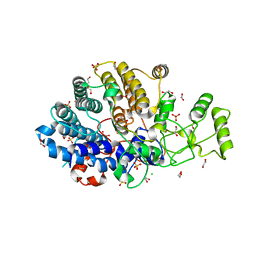 | | Bacteroidetes AC2a SusD-like | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koropatkin, N.M. | | Deposit date: | 2018-05-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | To Be Published
To Be Published
|
|
6RSJ
 
 | |
6Y6Z
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | Descriptor: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
8S7F
 
 | | Crystal structure of Escherichia coli LpxH in complex with EBL-2805 | | Descriptor: | MANGANESE (II) ION, N-[4-[4-[3,5-bis(chloranyl)phenyl]piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Mowbray, S.L, Bergfors, T, Jones, T.A. | | Deposit date: | 2024-03-01 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, synthesis, and in vitro biological evaluation of meta-sulfonamidobenzamide-based antibacterial LpxH inhibitors.
Eur.J.Med.Chem., 278, 2024
|
|
4RU4
 
 | | Crystal structure of the tailspike protein gp49 from Pseudomonas phage LKA1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Browning, C, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
5FWC
 
 | | Human Spectrin SH3 domain D48G, E7A, K60A | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, SPECTRIN ALPHA CHAIN, ... | | Authors: | Gallego, P, Navarro, S, Ventura, S, Reverter, D. | | Deposit date: | 2016-02-12 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human Spectrin SH3 Domain D48G, E7A, K60A
To be Published
|
|
8BY2
 
 | | Structure of the K+/H+ exchanger KefC with GSH. | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE MONOPHOSPHATE, GLUTATHIONE, ... | | Authors: | Gulati, A, Drew, D. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structure and mechanism of the K + /H + exchanger KefC.
Nat Commun, 15, 2024
|
|
8Q61
 
 | | Co-crystal structure of human AKT2 with compound 3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[4-(1-azanyl-3-methyl-3-oxidanyl-cyclobutyl)phenyl]-7-phenyl-1-propyl-pyrido[2,3-b][1,4]oxazin-2-one, GLYCEROL, ... | | Authors: | Harrison, T, Barker, O. | | Deposit date: | 2023-08-10 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification and development of a subtype-selective allosteric AKT inhibitor suitable for clinical development.
Sci Rep, 12, 2022
|
|
6Y8O
 
 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
