2O6I
 
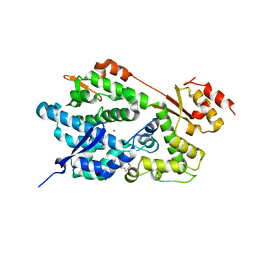 | | Structure of an Enterococcus Faecalis HD Domain Phosphohydrolase | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, HD domain protein, ... | | Authors: | Vorontsov, I.I, Minasov, G, Shuvalova, L, Brunzelle, J.S, Moy, S, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex.
J.Biol.Chem., 286, 2011
|
|
2O6K
 
 | | Crystal structure of UPF0346 from Staphylococcus aureus. Northeast Structural Genomics target ZR218. | | Descriptor: | UPF0346 protein MW1311 | | Authors: | Benach, J, Abashidze, M, Seetharaman, J, Wang, D, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UPF0346 from Staphylococcus aureus. Northeast Structural Genomics target ZR218.
To be Published
|
|
2O6L
 
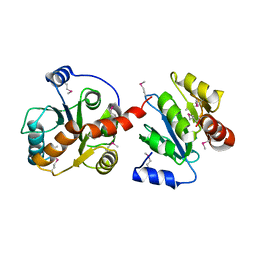 | |
2O6M
 
 | | H98Q mutant of the homing endonuclease I-PPOI complexed with DNA | | Descriptor: | 5'-D(*DTP*DTP*DGP*DAP*DCP*DTP*DCP*DTP*DCP*DTP*DTP*DAP*DAP*DGP*DAP*DGP*DAP*DGP*DTP*DCP*DA)-3', Intron-encoded endonuclease I-PpoI, MAGNESIUM ION, ... | | Authors: | Eastberg, J.H, Stoddard, B.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism.
Biochemistry, 46, 2007
|
|
2O6N
 
 | |
2O6P
 
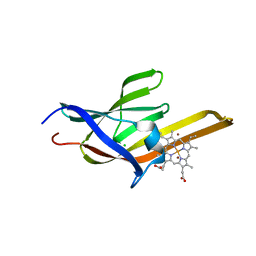 | | Crystal Structure of the heme-IsdC complex | | Descriptor: | CHLORIDE ION, Iron-regulated surface determinant protein C, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sharp, K.H, Schneider, S, Cockayne, A, Paoli, M. | | Deposit date: | 2006-12-08 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus.
J. Biol. Chem., 282, 2007
|
|
2O6Q
 
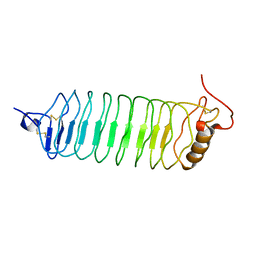 | |
2O6R
 
 | |
2O6S
 
 | |
2O6T
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (P2221). | | Descriptor: | CHLORIDE ION, THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O6U
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O6V
 
 | |
2O6W
 
 | | Crystal Structure of a Pentapeptide Repeat Protein (Rfr23) from the cyanobacterium Cyanothece 51142 | | Descriptor: | ARSENIC, Repeat Five Residue (Rfr) protein or pentapeptide repeat protein | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H, Pakrasi, H.B. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the structural variation between pentapeptide repeat proteins-Crystal structure of Rfr23 from Cyanothece 51142.
J.Struct.Biol., 162, 2008
|
|
2O6X
 
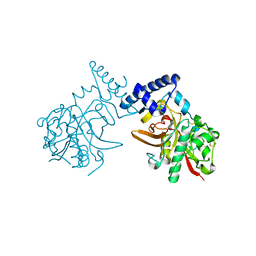 | | Crystal Structure of ProCathepsin L1 from Fasciola hepatica | | Descriptor: | Secreted cathepsin L 1 | | Authors: | Brinen, L.S, Dalton, J.P, Geiger, S, Marion, R, Stack, C.M. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional relationships in the virulence-associated cathepsin L proteases of the parasitic liver fluke, Fasciola hepatica.
J.Biol.Chem., 283, 2008
|
|
2O6Y
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides | | Descriptor: | Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-09 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
2O70
 
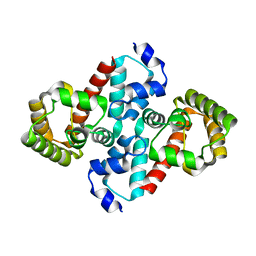 | | Structure of OHCU decarboxylase from zebrafish | | Descriptor: | OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-09 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O71
 
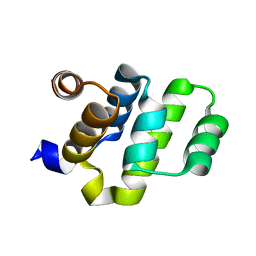 | | Crystal structure of RAIDD DD | | Descriptor: | Death domain-containing protein CRADD | | Authors: | Wu, H, Park, H. | | Deposit date: | 2006-12-09 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of RAIDD Death Domain Implicates Potential Mechanism of PIDDosome Assembly
J.Mol.Biol., 357, 2006
|
|
2O72
 
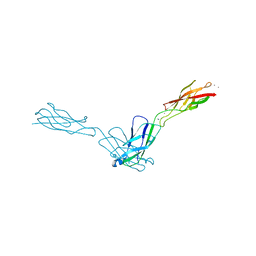 | | Crystal Structure Analysis of human E-cadherin (1-213) | | Descriptor: | CALCIUM ION, Epithelial-cadherin | | Authors: | Parisini, E, Wang, J.-H. | | Deposit date: | 2006-12-09 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human E-cadherin Domains 1 and 2, and Comparison with other Cadherins in the Context of Adhesion Mechanism
J.Mol.Biol., 373, 2007
|
|
2O73
 
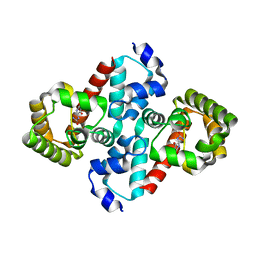 | | Structure of OHCU decarboxylase in complex with allantoin | | Descriptor: | 1-(2,5-DIOXO-2,5-DIHYDRO-1H-IMIDAZOL-4-YL)UREA, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O74
 
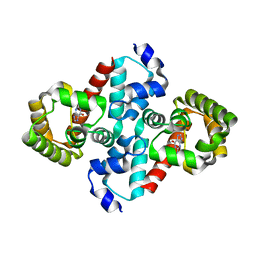 | | Structure of OHCU decarboxylase in complex with guanine | | Descriptor: | GUANINE, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O78
 
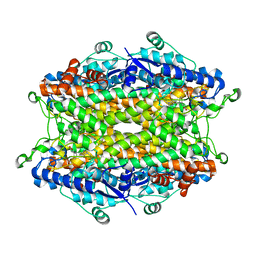 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides (His89Phe variant) complexed with cinnamic acid | | Descriptor: | PHENYLETHYLENECARBOXYLIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
2O79
 
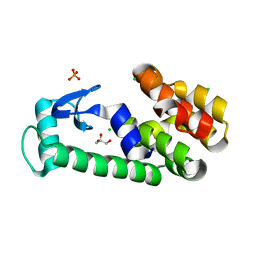 | | T4 lysozyme with C-terminal extension | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme, ... | | Authors: | Llinas, M, Crowder, S.M, Echols, N, Alber, T, Marqusee, S. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
2O7A
 
 | | T4 lysozyme C-terminal fragment | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme | | Authors: | Echols, N, Kwon, E, Marqusee, S.M, Alber, T. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
2O7B
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides, complexed with coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
2O7C
 
 | | Crystal structure of L-methionine-lyase from Pseudomonas | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2006-12-10 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the antitumour enzyme L-methionine gamma-lyase from Pseudomonas putida at 1.8 A resolution
J.Biochem.(Tokyo), 141, 2007
|
|
