7AKI
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxamide, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2020-10-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6G2I
 
 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) at 5.9 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1, Breast cancer type 1 susceptibility protein | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
6G2D
 
 | | Citrate-induced acetyl-CoA carboxylase (ACC-Cit) filament at 5.4 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
7A5M
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-17]-OMe | | Descriptor: | Ac-[2-Cl-F]-[ProM-2]-[ProM-17]-OMe, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6G2H
 
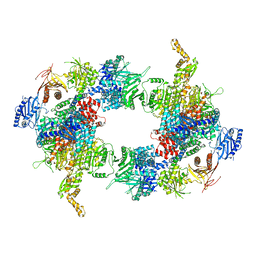 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) core at 4.6 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
3FCS
 
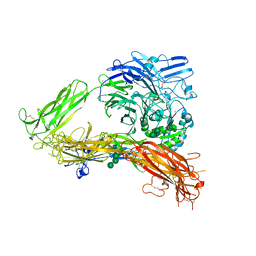 | | Structure of complete ectodomain of integrin aIIBb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
6SOY
 
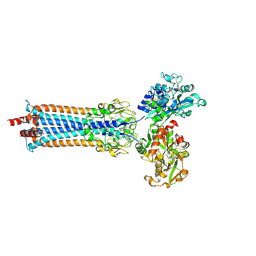 | | Trypanosoma brucei transferrin receptor in complex with human transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ESAG6, subunit of heterodimeric transferrin receptor, ... | | Authors: | Trevor, C, Carrington, M, Higgins, M.K. | | Deposit date: | 2019-08-30 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the trypanosome transferrin receptor reveals mechanisms of ligand recognition and immune evasion.
Nat Microbiol, 4, 2019
|
|
6SOZ
 
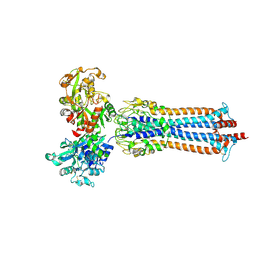 | | Glycosylated Trypanosoma brucei transferrin receptor in complex with human transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ESAG6, ... | | Authors: | Trevor, C, Carrington, M, Higgins, M.K. | | Deposit date: | 2019-08-30 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the trypanosome transferrin receptor reveals mechanisms of ligand recognition and immune evasion.
Nat Microbiol, 4, 2019
|
|
3FYQ
 
 | | Structure of Drosophila melanogaster talin IBS2 domain (residues 1981-2168) | | Descriptor: | CG6831-PA (Talin) | | Authors: | Cheung, T.Y.S, Fairchild, M.J, Zarivach, R, Tanentzapf, G, Van Petegem, F. | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the talin integrin binding domain 2.
J.Mol.Biol., 387, 2009
|
|
1QVI
 
 | | Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Angstrom resolution: flexibility and function in the head | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gourinath, S, Himmel, D.M, Brown, J.H, Reshetnikova, L, Szent-Gyrgyi, A.G, Cohen, C. | | Deposit date: | 2003-08-27 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of scallop Myosin s1 in the pre-power stroke state to 2.6 a resolution: flexibility and function in the head.
Structure, 11, 2003
|
|
3DYJ
 
 | |
2MYS
 
 | |
4OQS
 
 | | Crystal structure of CYP105AS1 | | Descriptor: | CYP105AS1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2014-02-10 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4EHU
 
 | |
4EIA
 
 | |
4EHT
 
 | |
3L4F
 
 | | Crystal Structure of betaPIX Coiled-Coil Domain and Shank PDZ Complex | | Descriptor: | Rho guanine nucleotide exchange factor 7, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Im, Y.J, Kang, G.B, Lee, J.H, Song, H.E, Park, K.R, Kim, E, Song, W.K, Park, D, Eom, S.H. | | Deposit date: | 2009-12-19 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for asymmetric association of the betaPIX coiled coil and shank PDZ
J.Mol.Biol., 397, 2010
|
|
4CC3
 
 | | Complex of human Tuba C-terminal SH3 domain and Mena proline-rich peptide - H3 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CHLORIDE ION, DYNAMIN-BINDING PROTEIN, ... | | Authors: | Polle, L, Rigano, L, Julian, R, Ireton, K, Schubert, W.-D. | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Details of Human Tuba Recruitment by Inlc of Listeria Monocytogenes Elucidate Bacterial Cell-Cell Spreading.
Structure, 22, 2014
|
|
4CC2
 
 | | Complex of human Tuba C-terminal SH3 domain with human N-WASP proline- rich peptide - P212121 | | Descriptor: | CHLORIDE ION, DYNAMIN-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Polle, L, Rigano, L, Julian, R, Ireton, K, Schubert, W.-D. | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Human Tuba Recruitment by Inlc of Listeria Monocytogenes Elucidate Bacterial Cell-Cell Spreading.
Structure, 22, 2014
|
|
4CC7
 
 | | Crystal structure of the sixth or C-terminal SH3 domain of human Tuba in complex with proline-rich peptides of N-WASP. Space group P41 | | Descriptor: | CHLORIDE ION, DYNAMIN-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Polle, L, Rigano, L, Julian, R, Ireton, K, Schubert, W.-D. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Details of Human Tuba Recruitment by Inlc of Listeria Monocytogenes Elucidate Bacterial Cell-Cell Spreading.
Structure, 22, 2014
|
|
6A5Q
 
 | |
6A5S
 
 | | Structure of 14-3-3 gamma in complex with TFEB 14-3-3 binding motif | | Descriptor: | 14-3-3 protein gamma, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Xu, Y, Ren, J.Q, Feng, W. | | Deposit date: | 2018-06-25 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | YWHA/14-3-3 proteins recognize phosphorylated TFEB by a noncanonical mode for controlling TFEB cytoplasmic localization.
Autophagy, 15, 2019
|
|
5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | Authors: | Yu, X, Jih, J, Jiang, J, Zhou, H. | | Deposit date: | 2017-04-24 | | Release date: | 2017-06-28 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
3VU7
 
 | | Crystal structure of REV1-REV7-REV3 ternary complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
7MON
 
 | | Structure of human RIPK3-MLKL complex | | Descriptor: | Mixed lineage kinase domain-like protein, N-[4-({2-[(cyclopropanecarbonyl)amino]pyridin-4-yl}oxy)-3-fluorophenyl]-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Meng, Y, Davies, K.A, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2021-05-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
